Python for Data Analysis
Overview
Teaching: 150 min
Exercises: 30 minQuestions
How can I summarize my data in Python?
How can Python help make my research more reproducible?
How can I combine two datasets from different sources?
How can data tidying facilitate answering analysis questions?
Objectives
To become familiar with the common methods of the Python pandas library.
To be able to use pandas to prepare data for analysis.
To be able to combine two different data sources using joins.
To be able to create plots and summary tables to answer analysis questions.
Contents
- Getting started
- An introduction to data analysis with pandas
- Cleaning up data
- Joining data frames
- Analyzing combined data
- Finishing with Git and GitHub
- Bonus exercises
Getting Started
Yesterday we spent a lot of time making plots in Python using the seaborn library. Visualizing data using plots is a very powerful skill in Python, but what if we would like to work with only a subset of our data? Or clean up messy data, calculate summary statistics, create a new variable, or join two datasets together? There are several different methods for doing this in Python, and we will touch on a few today using the fast and powerful pandas library.
- First, navigate to the
un-reportsdirectory in your Command Line Interface (i.e., Anaconda Prompt for Windows and Terminal for MacOS and Linux) -
Launch JupyterLab.
jupyter lab - Once JupyterLab is opened in your web browser, you can check you are at the correct directory by seeing if the JupyterLab File Browser (located on the left side panel) shows the folders and files inside the
un-reportsdirectory. - Create a new Jupyter notebook file for our work.
- Make sure you are at the
un-reportshome directory. - On the Launcher tab (the main window on the right) click “Python 3” under the Notebook category.
- Make sure you are at the
- Then you should see a new file named
Untitled.ipynbbeen created on the File Browser. Left-click the file on the File Browser and rename it togapminder_data_analysis.ipynb.
Reading in the data
We will start by reading in the complete gapminder dataset that we used yesterday into our fresh new Jupyter notebook.
Let’s type the code into a cell: gapminder = pd.read_csv("./data/gapminder_data.csv")
Exercise
If we look in the console now, we’ll see we’ve received an error message saying that “name ‘pd’ is not defined”. Hint: Libraries…
Solution
What this means is that Python did not recognize the
pdpart of the code and thus cannot find theread_csv()function we are trying to call. The reason for this usually is that we are trying to run a function from a library that we have not yet imported. This is a very common error message that you will probably see again when using Python. It’s important to remember that you will need to import any libraries you want to use into Python each time you start a new notebook. Theread_csvfunction comes from the pandas library so we will just import the pandas library and run the code again.
Now that we know what’s wrong, We will use the read_csv() function from the pandas library.
Import the pandas library (along with another common library numpy) and read in the gapminder dataset using the code below.
import numpy as np
import pandas as pd
gapminder = pd.read_csv("./data/gapminder_data.csv")
gapminder # this line is just to show the data in the Jupyter notebook output
country year pop continent lifeExp gdpPercap
0 Afghanistan 1952 8425333.0 Asia 28.801 779.445314
1 Afghanistan 1957 9240934.0 Asia 30.332 820.853030
2 Afghanistan 1962 10267083.0 Asia 31.997 853.100710
3 Afghanistan 1967 11537966.0 Asia 34.020 836.197138
4 Afghanistan 1972 13079460.0 Asia 36.088 739.981106
... ... ... ... ... ... ...
1699 Zimbabwe 1987 9216418.0 Africa 62.351 706.157306
1700 Zimbabwe 1992 10704340.0 Africa 60.377 693.420786
1701 Zimbabwe 1997 11404948.0 Africa 46.809 792.449960
1702 Zimbabwe 2002 11926563.0 Africa 39.989 672.038623
1703 Zimbabwe 2007 12311143.0 Africa 43.487 469.709298
[1704 rows x 6 columns]
The output above gives us an overview of the data with its first and last few rows, the names of the columns, and the numbers of rows and columns.
If we want more information, we can apply the info() method to a data frame to print some basic information about it.
In Python we use the dot notation to apply a method to an object.
Note: When applying a method, we always need to follow the method name by a pair of parentheses, even if we are not passing any arguments to the method.
gapminder.info()
<class 'pandas.core.frame.DataFrame'>
RangeIndex: 1704 entries, 0 to 1703
Data columns (total 6 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 country 1704 non-null object
1 year 1704 non-null int64
2 pop 1704 non-null float64
3 continent 1704 non-null object
4 lifeExp 1704 non-null float64
5 gdpPercap 1704 non-null float64
dtypes: float64(3), int64(1), object(2)
memory usage: 80.0+ KB
- The first line in the output indicates
gapminder, the output from thepd.read_csv()function that we called earlier, is a pandas DataFrame. A pandas DataFrame is a two-dimensional rectangular table of data with rows and columns. It is the main data structure that we will be dealing with when working with pandas. - The information also shows the data type (“Dtype”) of each column.
- Not surprisingly, the
yearcolumn has an integer data type (“int64”). - A few other columns have the data type of floating number (“float64”).
- The
countrycolumn has a data type of “object”, indicating the data is a string or mixed type.
- Not surprisingly, the
Sometimes (especially when our data has many rows) we just want to take a look at the first few rows of the data. We can apply the head() method to select the first few rows of a data frame.
gapminder.head()
country year pop continent lifeExp gdpPercap
0 Afghanistan 1952 8425333.0 Asia 28.801 779.445314
1 Afghanistan 1957 9240934.0 Asia 30.332 820.853030
2 Afghanistan 1962 10267083.0 Asia 31.997 853.100710
3 Afghanistan 1967 11537966.0 Asia 34.020 836.197138
4 Afghanistan 1972 13079460.0 Asia 36.088 739.981106
By default, the head() method selects the first 5 rows of the data frame. You can change the number of rows by passing a number as an argument to the method.
For example, we can use the code below to select the first 3 rows.
gapminder.head(3)
country year pop continent lifeExp gdpPercap
0 Afghanistan 1952 8425333.0 Asia 28.801 779.445314
1 Afghanistan 1957 9240934.0 Asia 30.332 820.853030
2 Afghanistan 1962 10267083.0 Asia 31.997 853.100710
Similarly, we can apply the tail() method to select the last few rows of a data frame.
gapminder.tail()
country year pop continent lifeExp gdpPercap
1699 Zimbabwe 1987 9216418.0 Africa 62.351 706.157306
1700 Zimbabwe 1992 10704340.0 Africa 60.377 693.420786
1701 Zimbabwe 1997 11404948.0 Africa 46.809 792.449960
1702 Zimbabwe 2002 11926563.0 Africa 39.989 672.038623
1703 Zimbabwe 2007 12311143.0 Africa 43.487 469.709298
Now we have the tools necessary to work through this lesson.
An introduction to data analysis with pandas
Get stats fast with describe()
Pandas has a handy method describe() that will generate the summary statistics of the data.
gapminder.describe()
year pop lifeExp gdpPercap
count 1704.00000 1.704000e+03 1704.000000 1704.000000
mean 1979.50000 2.960121e+07 59.474439 7215.327081
std 17.26533 1.061579e+08 12.917107 9857.454543
min 1952.00000 6.001100e+04 23.599000 241.165876
25% 1965.75000 2.793664e+06 48.198000 1202.060309
50% 1979.50000 7.023596e+06 60.712500 3531.846988
75% 1993.25000 1.958522e+07 70.845500 9325.462346
max 2007.00000 1.318683e+09 82.603000 113523.132900
The output above shows the summary (or descriptive) statistics for the four numerical columns in our data.
If we are interested in specific columns with specific statistics, we can also apply the agg() method to aggregate a column based on some aggregation functions.
Let’s say we would like to know what is the mean life expectancy in the dataset.
(
gapminder
.agg({'lifeExp' : 'mean'})
)
lifeExp 59.474439
dtype: float64
Other aggregation functions for common descriptive statistics include median, min, max, std (for standard deviation), and var (for variance).
Narrow down rows with query()
Let’s take a look at the value we just calculated, which tells us the mean life expectancy for all rows in the data was 59.47. That seems a bit low, doesn’t it? What’s going on?
Well, remember the dataset contains rows from many different years and many different countries. It’s likely that life expectancy has increased overtime, so it may not make sense to average over all the years at the same time.
Use the max() method to find the most recent year in the data set.
Practice getting descriptive statistics
Find the most recent year in the dataset.
Solution:
( gapminder['year'] .max() )2007
So we see that the most recent year in the dataset is 2007.
Let’s calculate the life expectancy for all countries for only that year.
To do that, we will apply the query() method to only use the rows for that year before calculating the mean life expectancy.
(
gapminder
.query("year == 2007")
.agg({'lifeExp' : 'mean'})
)
lifeExp 67.007423
dtype: float64
Querying the dataset
What is the mean GDP per capita for the first year in the dataset? Hint: The data frame has a column called “gdpPercap”.
Solution
Identify the earliest year in our dataset by applying the
aggmethod.( gapminder .agg({'year' : 'min'}) )year 1952 dtype: int64We see here that the first year in the dataset is 1952. Query the data to only include year 1952, and determine the mean GDP per capita.
( gapminder .query("year == 1952") .agg({'gdpPercap' : 'mean'}) )gdpPercap 3725.276046 dtype: float64By chaining the two methods
query()andagg()we were able to calculate the mean GDP per capita in the year 1952.
Notice how the method chaining allows us to combine these two simple steps into a more complicated data extraction?
We took the data, queried the year column, selected the gdpPercap columns, then took its mean value.
The string argument we passed to query() needs to be an expression that will return TRUE or FALSE for each row.
We use == (double equals) when evaluating if two values are equal, and we use = (single equal) when assigning values.
Try changing the code above to use query("year = 2007") and see what happens.
Other common Python comparison operators
>greater than<less than>=greater than or equal to<=less than or equal to==equal!=not equal
We can also use the operator == to evaluate if two strings are the same.
For example, the code below returns all the rows from the United States.
(
gapminder
.query("country == 'United States'")
)
country year pop continent lifeExp gdpPercap
1608 United States 1952 157553000.0 Americas 68.440 13990.48208
1609 United States 1957 171984000.0 Americas 69.490 14847.12712
1610 United States 1962 186538000.0 Americas 70.210 16173.14586
1611 United States 1967 198712000.0 Americas 70.760 19530.36557
1612 United States 1972 209896000.0 Americas 71.340 21806.03594
1613 United States 1977 220239000.0 Americas 73.380 24072.63213
1614 United States 1982 232187835.0 Americas 74.650 25009.55914
1615 United States 1987 242803533.0 Americas 75.020 29884.35041
1616 United States 1992 256894189.0 Americas 76.090 32003.93224
1617 United States 1997 272911760.0 Americas 76.810 35767.43303
1618 United States 2002 287675526.0 Americas 77.310 39097.09955
1619 United States 2007 301139947.0 Americas 78.242 42951.65309
Note: In a
query()expression, any string values (e.g., United States in the code above) need to be wrapped with quotation marks.
Note: In a
query()expression, any column names that does not include any special characters (e.g., a white space) do not need to be wrapped with anything. However, if a column name does include special characters, the name needs to be wrapped with a pair of backticks``(the key above the Tab key on your keyboard).
Oftentimes we may wish to query the data based on more than a single criterion.
In a query() expression we can combine multiple criteria with Python logical operators and or or.
For example, the code below returns all the rows that are from the United States and after 2000.
(
gapminder
.query("country == 'United States' and year > 2000")
)
country year pop continent lifeExp gdpPercap
1618 United States 2002 287675526.0 Americas 77.310 39097.09955
1619 United States 2007 301139947.0 Americas 78.242 42951.65309
Note if the logical operators are all and, we can also separate them by chaining multiple query() methods.
The code below generates the same results as above.
(
gapminder
.query("country == 'United States'")
.query("year > 2000")
)
Sometimes we may wish to query the data based on whether a value is from a list or not.
In a query() expression we can use the Python membership operator in to achieve that.
For example, the code below returns all the rows from a list of countries (the United States and Canada).
(
gapminder
.query("country in ['United States', 'Canada']")
)
country year pop continent lifeExp gdpPercap
240 Canada 1952 14785584.0 Americas 68.750 11367.16112
241 Canada 1957 17010154.0 Americas 69.960 12489.95006
242 Canada 1962 18985849.0 Americas 71.300 13462.48555
243 Canada 1967 20819767.0 Americas 72.130 16076.58803
244 Canada 1972 22284500.0 Americas 72.880 18970.57086
245 Canada 1977 23796400.0 Americas 74.210 22090.88306
246 Canada 1982 25201900.0 Americas 75.760 22898.79214
247 Canada 1987 26549700.0 Americas 76.860 26626.51503
248 Canada 1992 28523502.0 Americas 77.950 26342.88426
249 Canada 1997 30305843.0 Americas 78.610 28954.92589
250 Canada 2002 31902268.0 Americas 79.770 33328.96507
251 Canada 2007 33390141.0 Americas 80.653 36319.23501
1608 United States 1952 157553000.0 Americas 68.440 13990.48208
1609 United States 1957 171984000.0 Americas 69.490 14847.12712
1610 United States 1962 186538000.0 Americas 70.210 16173.14586
1611 United States 1967 198712000.0 Americas 70.760 19530.36557
1612 United States 1972 209896000.0 Americas 71.340 21806.03594
1613 United States 1977 220239000.0 Americas 73.380 24072.63213
1614 United States 1982 232187835.0 Americas 74.650 25009.55914
1615 United States 1987 242803533.0 Americas 75.020 29884.35041
1616 United States 1992 256894189.0 Americas 76.090 32003.93224
1617 United States 1997 272911760.0 Americas 76.810 35767.43303
1618 United States 2002 287675526.0 Americas 77.310 39097.09955
1619 United States 2007 301139947.0 Americas 78.242 42951.65309
In a query() expression we can refer to variables in the environment by prefixing them with an ‘@’ character. For example, the code below generates the same results as above.
country_list = ['United States', 'Canada']
(
gapminder
.query("country in @country_list")
)
Lastly, we can use the not in operator to evaluate if a value is not in a list.
For example, the code below returns all the rows for 2007 in the Americas except for the United States and Canada.
(
gapminder
.query("year == 2007")
.query("continent == 'Americas'")
.query("country not in ['United States', 'Canada']")
)
country year pop continent lifeExp gdpPercap
59 Argentina 2007 40301927.0 Americas 75.320 12779.379640
143 Bolivia 2007 9119152.0 Americas 65.554 3822.137084
179 Brazil 2007 190010647.0 Americas 72.390 9065.800825
287 Chile 2007 16284741.0 Americas 78.553 13171.638850
311 Colombia 2007 44227550.0 Americas 72.889 7006.580419
359 Costa Rica 2007 4133884.0 Americas 78.782 9645.061420
395 Cuba 2007 11416987.0 Americas 78.273 8948.102923
443 Dominican Republic 2007 9319622.0 Americas 72.235 6025.374752
455 Ecuador 2007 13755680.0 Americas 74.994 6873.262326
479 El Salvador 2007 6939688.0 Americas 71.878 5728.353514
611 Guatemala 2007 12572928.0 Americas 70.259 5186.050003
647 Haiti 2007 8502814.0 Americas 60.916 1201.637154
659 Honduras 2007 7483763.0 Americas 70.198 3548.330846
791 Jamaica 2007 2780132.0 Americas 72.567 7320.880262
995 Mexico 2007 108700891.0 Americas 76.195 11977.574960
1115 Nicaragua 2007 5675356.0 Americas 72.899 2749.320965
1187 Panama 2007 3242173.0 Americas 75.537 9809.185636
1199 Paraguay 2007 6667147.0 Americas 71.752 4172.838464
1211 Peru 2007 28674757.0 Americas 71.421 7408.905561
1259 Puerto Rico 2007 3942491.0 Americas 78.746 19328.709010
1559 Trinidad and Tobago 2007 1056608.0 Americas 69.819 18008.509240
1631 Uruguay 2007 3447496.0 Americas 76.384 10611.462990
1643 Venezuela 2007 26084662.0 Americas 73.747 11415.805690
Grouping rows using groupby()
We see that the life expectancy in 2007 is much larger than the value we got using all of the rows.
It seems life expectancy is increasing which is good news.
But now we might be interested in calculating the mean for each year.
Rather than doing a bunch of different query()’s for each year, we can instead use the groupby() method.
This method allows us to tell the code to treat the rows in logical groups, so rather than aggregating over all the rows, we will get one summary value for each group.
“Group by” is often often referred to as split-apply-combine.
(
gapminder
.groupby('year')
)
<pandas.core.groupby.generic.DataFrameGroupBy object at 0x157f8d2b0>
If we just apply the groupby() method to our data frame, we would only get a data object called “DataFrameGroupBy”.
This is because this method only group the data based on our specification, in this case, by year.
We have not specify what kind of the aggregation functions that we wish to apply to each of the groups.
We can take a closer look at the data object after the groupby() by its indices property.
(
gapminder
.groupby('year')
.indices
)
{1952: array([ 0, 12, 24, 36, 48, 60, 72, 84, 96, 108, 120,
132, 144, 156, 168, 180, 192, 204, 216, 228, 240, 252,
264, 276, 288, 300, 312, 324, 336, 348, 360, 372, 384,
396, 408, 420, 432, 444, 456, 468, 480, 492, 504, 516,
528, 540, 552, 564, 576, 588, 600, 612, 624, 636, 648,
660, 672, 684, 696, 708, 720, 732, 744, 756, 768, 780,
792, 804, 816, 828, 840, 852, 864, 876, 888, 900, 912,
924, 936, 948, 960, 972, 984, 996, 1008, 1020, 1032, 1044,
1056, 1068, 1080, 1092, 1104, 1116, 1128, 1140, 1152, 1164, 1176,
1188, 1200, 1212, 1224, 1236, 1248, 1260, 1272, 1284, 1296, 1308,
1320, 1332, 1344, 1356, 1368, 1380, 1392, 1404, 1416, 1428, 1440,
1452, 1464, 1476, 1488, 1500, 1512, 1524, 1536, 1548, 1560, 1572,
1584, 1596, 1608, 1620, 1632, 1644, 1656, 1668, 1680, 1692]),
...
2007: array([ 11, 23, 35, 47, 59, 71, 83, 95, 107, 119, 131,
143, 155, 167, 179, 191, 203, 215, 227, 239, 251, 263,
275, 287, 299, 311, 323, 335, 347, 359, 371, 383, 395,
407, 419, 431, 443, 455, 467, 479, 491, 503, 515, 527,
539, 551, 563, 575, 587, 599, 611, 623, 635, 647, 659,
671, 683, 695, 707, 719, 731, 743, 755, 767, 779, 791,
803, 815, 827, 839, 851, 863, 875, 887, 899, 911, 923,
935, 947, 959, 971, 983, 995, 1007, 1019, 1031, 1043, 1055,
1067, 1079, 1091, 1103, 1115, 1127, 1139, 1151, 1163, 1175, 1187,
1199, 1211, 1223, 1235, 1247, 1259, 1271, 1283, 1295, 1307, 1319,
1331, 1343, 1355, 1367, 1379, 1391, 1403, 1415, 1427, 1439, 1451,
1463, 1475, 1487, 1499, 1511, 1523, 1535, 1547, 1559, 1571, 1583,
1595, 1607, 1619, 1631, 1643, 1655, 1667, 1679, 1691, 1703])}
It shows the indices of each group (i.e., each year in this case).
We can double-check the indices of the first group by manually querying the data for year 1952. The first column in the output shows the indices.
(
gapminder
.query("year == 1952")
)
country year pop continent lifeExp gdpPercap
0 Afghanistan 1952 8425333.0 Asia 28.801 779.445314
12 Albania 1952 1282697.0 Europe 55.230 1601.056136
24 Algeria 1952 9279525.0 Africa 43.077 2449.008185
36 Angola 1952 4232095.0 Africa 30.015 3520.610273
48 Argentina 1952 17876956.0 Americas 62.485 5911.315053
... ... ... ... ... ... ...
1644 Vietnam 1952 26246839.0 Asia 40.412 605.066492
1656 West Bank and Gaza 1952 1030585.0 Asia 43.160 1515.592329
1668 Yemen Rep. 1952 4963829.0 Asia 32.548 781.717576
1680 Zambia 1952 2672000.0 Africa 42.038 1147.388831
1692 Zimbabwe 1952 3080907.0 Africa 48.451 406.884115
In practice, we will just trust the groupby() method will do its job, and just apply aggregation functions of our interest by calling the aggregation method agg() after the groupby().
(
gapminder
.groupby('year')
.agg({'lifeExp' : 'mean'})
)
lifeExp
year
1952 49.057620
1957 51.507401
1962 53.609249
1967 55.678290
1972 57.647386
1977 59.570157
1982 61.533197
1987 63.212613
1992 64.160338
1997 65.014676
2002 65.694923
2007 67.007423
The groupby() method expects you to pass in the name of a column (or a list of columns) in your data.
Grouping the data
Try calculating the mean life expectancy by continent.
Solution
( gapminder .groupby('continent') .agg({'lifeExp' : 'mean'}) )lifeExp continent Africa 48.865330 Americas 64.658737 Asia 60.064903 Europe 71.903686 Oceania 74.326208By chaining the two methods
groupby()andagg()we are able to calculate the mean life expectancy by continent.
Sometimes we may wish to apply more than one aggregation method.
For example, we may want to know the mean and minimum life expectancy by continents.
To do so, we can use the aggregation method called agg() and pass it a list of aggregation methods.
(
gapminder
.groupby('continent')
.agg({'lifeExp' : ['mean', 'min']})
)
lifeExp
mean min
continent
Africa 48.865330 23.599
Americas 64.658737 37.579
Asia 60.064903 28.801
Europe 71.903686 43.585
Oceania 74.326208 69.120
Sort data with sort_values()
The sort_values() method allows us to sort our data by some value.
Let’s use the full gapminder data.
We will take the mean value for each continent in 2007 and then sort it so the continents with the longest life expectancy are on top.
Which continent might you guess has be the highest life expectancy before running the code?
(
gapminder
.query("year == 2007")
.groupby('continent')
.agg({'lifeExp' : 'mean'})
.sort_values('lifeExp', ascending=False)
)
lifeExp
continent
Oceania 80.719500
Europe 77.648600
Americas 73.608120
Asia 70.728485
Africa 54.806038
Notice we passed the argument ascending=False to the sort_values() method to sort the values in a descending order so the largest values are on top.
The default is to put the smallest values on top.
Make new variables with assign()
Sometimes we want to create a new column in our data.
We can use the pandas assign() method to assign new columns to a data frame.
We have a column for the population and the GDP per capita. If we wanted to get the total GDP, we could multiply the per capita GDP values by the total population. Below is what the code would look like:
Here we use the lambda function.
(
gapminder
.assign(gdp=lambda df: df['pop'] * df['gdpPercap'])
)
country year pop continent lifeExp gdpPercap gdp
0 Afghanistan 1952 8425333.0 Asia 28.801 779.445314 6.567086e+09
1 Afghanistan 1957 9240934.0 Asia 30.332 820.853030 7.585449e+09
2 Afghanistan 1962 10267083.0 Asia 31.997 853.100710 8.758856e+09
3 Afghanistan 1967 11537966.0 Asia 34.020 836.197138 9.648014e+09
4 Afghanistan 1972 13079460.0 Asia 36.088 739.981106 9.678553e+09
... ... ... ... ... ... ... ...
1699 Zimbabwe 1987 9216418.0 Africa 62.351 706.157306 6.508241e+09
1700 Zimbabwe 1992 10704340.0 Africa 60.377 693.420786 7.422612e+09
1701 Zimbabwe 1997 11404948.0 Africa 46.809 792.449960 9.037851e+09
1702 Zimbabwe 2002 11926563.0 Africa 39.989 672.038623 8.015111e+09
1703 Zimbabwe 2007 12311143.0 Africa 43.487 469.709298 5.782658e+09
[1704 rows x 7 columns]
This will add a new column called “gdp” to our data. We use the column names as if they were regular values that we want to perform mathematical operations on and provide the name in front of an equals sign.
Assigning multiple columns
We can also assign multiple columns by separating them with a comma inside
assign(). Try making a new column for this data frame called popInMillions that is the population in million.Solution:
( gapminder .assign(gdp=lambda df: df['pop'] * df['gdpPercap'], popInMillions=lambda df: df['pop'] / 1_000_000) )country year pop continent lifeExp gdpPercap gdp popInMillions 0 Afghanistan 1952 8425333.0 Asia 28.801 779.445314 6.567086e+09 8.425333 1 Afghanistan 1957 9240934.0 Asia 30.332 820.853030 7.585449e+09 9.240934 2 Afghanistan 1962 10267083.0 Asia 31.997 853.100710 8.758856e+09 10.267083 3 Afghanistan 1967 11537966.0 Asia 34.020 836.197138 9.648014e+09 11.537966 4 Afghanistan 1972 13079460.0 Asia 36.088 739.981106 9.678553e+09 13.079460 ... ... ... ... ... ... ... ... ... 1699 Zimbabwe 1987 9216418.0 Africa 62.351 706.157306 6.508241e+09 9.216418 1700 Zimbabwe 1992 10704340.0 Africa 60.377 693.420786 7.422612e+09 10.704340 1701 Zimbabwe 1997 11404948.0 Africa 46.809 792.449960 9.037851e+09 11.404948 1702 Zimbabwe 2002 11926563.0 Africa 39.989 672.038623 8.015111e+09 11.926563 1703 Zimbabwe 2007 12311143.0 Africa 43.487 469.709298 5.782658e+09 12.311143 [1704 rows x 8 columns]
Subset columns
Sometimes we may want to select a subset of columns from our data based on the column names.
If we want to select a single column, we can use the square bracket [] notation.
For example, if we want to select the population column from our data, we can do:
gapminder['pop']
0 8425333.0
1 9240934.0
2 10267083.0
3 11537966.0
4 13079460.0
...
1699 9216418.0
1700 10704340.0
1701 11404948.0
1702 11926563.0
1703 12311143.0
Name: pop, Length: 1704, dtype: float64
If we want to select multiple columns, we can pass a list of column names into (another) pair of square brackets.
gapminder[['pop', 'year']]
pop year
0 8425333.0 1952
1 9240934.0 1957
2 10267083.0 1962
3 11537966.0 1967
4 13079460.0 1972
... ... ...
1699 9216418.0 1987
1700 10704340.0 1992
1701 11404948.0 1997
1702 11926563.0 2002
1703 12311143.0 2007
[1704 rows x 2 columns]
Note: There are two nested pairs of square brackets in the code above. The outer square brackets is the notation for selecting columns from a data frame by name. The inner square brackets define a Python list that contains the column names. Try removing one pair of brackets and see what happens.
Another way to select columns is to use the filter() method.
The code below gives the same output as the above.
(
gapminder
.filter(['pop', 'year'])
)
We can also apply the drop() method to drop/remove particular columns.
For example, if we want everything but the continent and population columns, we can do:
(
gapminder
.drop(columns=['continent', 'pop'])
)
country year lifeExp gdpPercap
0 Afghanistan 1952 28.801 779.445314
1 Afghanistan 1957 30.332 820.853030
2 Afghanistan 1962 31.997 853.100710
3 Afghanistan 1967 34.020 836.197138
4 Afghanistan 1972 36.088 739.981106
... ... ... ... ...
1699 Zimbabwe 1987 62.351 706.157306
1700 Zimbabwe 1992 60.377 693.420786
1701 Zimbabwe 1997 46.809 792.449960
1702 Zimbabwe 2002 39.989 672.038623
1703 Zimbabwe 2007 43.487 469.709298
[1704 rows x 4 columns]
selecting columns
Create a data frame with only the
country,continent,year, andlifeExpcolumns.Solution:
There are multiple ways to do this exercise. Here are two different possibilities.
( gapminder .filter(['country', 'continent', 'year', 'lifeExp']) )country continent year lifeExp 0 Afghanistan Asia 1952 28.801 1 Afghanistan Asia 1957 30.332 2 Afghanistan Asia 1962 31.997 3 Afghanistan Asia 1967 34.020 4 Afghanistan Asia 1972 36.088 ... ... ... ... ... 1699 Zimbabwe Africa 1987 62.351 1700 Zimbabwe Africa 1992 60.377 1701 Zimbabwe Africa 1997 46.809 1702 Zimbabwe Africa 2002 39.989 1703 Zimbabwe Africa 2007 43.487 [1704 rows x 4 columns]( gapminder .drop(columns=['pop', 'gdpPercap']) )country year continent lifeExp 0 Afghanistan 1952 Asia 28.801 1 Afghanistan 1957 Asia 30.332 2 Afghanistan 1962 Asia 31.997 3 Afghanistan 1967 Asia 34.020 4 Afghanistan 1972 Asia 36.088 ... ... ... ... ... 1699 Zimbabwe 1987 Africa 62.351 1700 Zimbabwe 1992 Africa 60.377 1701 Zimbabwe 1997 Africa 46.809 1702 Zimbabwe 2002 Africa 39.989 1703 Zimbabwe 2007 Africa 43.487 [1704 rows x 4 columns]
Bonus: Additional features of the
filter()methodThe
filter()method can be used to filter columns by their names. It may become handy if you are working with a dataset that has a lot of columns. For example, let’s say we wanted to select the year column and all the columns that contain the letter “e”. You can do that with:( gapminder .filter(like='e') )year continent lifeExp gdpPercap 0 1952 Asia 28.801 779.445314 1 1957 Asia 30.332 820.853030 2 1962 Asia 31.997 853.100710 3 1967 Asia 34.020 836.197138 4 1972 Asia 36.088 739.981106 ... ... ... ... ... 1699 1987 Africa 62.351 706.157306 1700 1992 Africa 60.377 693.420786 1701 1997 Africa 46.809 792.449960 1702 2002 Africa 39.989 672.038623 1703 2007 Africa 43.487 469.709298 [1704 rows x 4 columns]This returns the four columns we are interested in.
Applying
filter()with regular expressionFor those of you who know regular expression (pattern matching in text), the
filter()method also supports it. For example, let’s say we want to select all the columns that start with the letter “c”. We can do that with:Solution
( gapminder .filter(regex='^c') )country continent 0 Afghanistan Asia 1 Afghanistan Asia 2 Afghanistan Asia 3 Afghanistan Asia 4 Afghanistan Asia ... ... ... 1699 Zimbabwe Africa 1700 Zimbabwe Africa 1701 Zimbabwe Africa 1702 Zimbabwe Africa 1703 Zimbabwe Africa [1704 rows x 2 columns]Similarly, if we want to select all the columns that end with the letter “p”. We can do that with:
Solution
( gapminder .filter(regex='p$') )pop lifeExp gdpPercap 0 8425333.0 28.801 779.445314 1 9240934.0 30.332 820.853030 2 10267083.0 31.997 853.100710 3 11537966.0 34.020 836.197138 4 13079460.0 36.088 739.981106 ... ... ... ... 1699 9216418.0 62.351 706.157306 1700 10704340.0 60.377 693.420786 1701 11404948.0 46.809 792.449960 1702 11926563.0 39.989 672.038623 1703 12311143.0 43.487 469.709298 [1704 rows x 3 columns]
Changing the shape of the data
Data comes in many shapes and sizes, and one way we classify data is either “wide” or “long.” Data that is “long” has one row per observation. The gapminder data is in a long format. We have one row for each country for each year and each different measurement for that country is in a different column. We might describe this data as “tidy” because it makes it easy to work with pandas and seaborn. As tidy as it may be, sometimes we may want our data in a “wide” format. Typically in “wide” format each row represents a group of observations and each value is placed in a different column rather than a different row. For example maybe we want only one row per country and want to spread the life expectancy values into different columns (one for each year).
The pandas methods pivot() and melt() make it easy to switch between the two formats.
(
gapminder
.filter(['country', 'continent', 'year', 'lifeExp'])
.pivot(columns='year',
index=['country', 'continent'],
values='lifeExp')
)
year 1952 1957 1962 1967 1972 1977 1982 1987 1992 1997 2002 2007
country continent
Afghanistan Asia 28.801 30.332 31.997 34.020 36.088 38.438 39.854 40.822 41.674 41.763 42.129 43.828
Albania Europe 55.230 59.280 64.820 66.220 67.690 68.930 70.420 72.000 71.581 72.950 75.651 76.423
Algeria Africa 43.077 45.685 48.303 51.407 54.518 58.014 61.368 65.799 67.744 69.152 70.994 72.301
Angola Africa 30.015 31.999 34.000 35.985 37.928 39.483 39.942 39.906 40.647 40.963 41.003 42.731
Argentina Americas 62.485 64.399 65.142 65.634 67.065 68.481 69.942 70.774 71.868 73.275 74.340 75.320
... ... ... ... ... ... ... ... ... ... ... ... ...
Vietnam Asia 40.412 42.887 45.363 47.838 50.254 55.764 58.816 62.820 67.662 70.672 73.017 74.249
West Bank and Gaza Asia 43.160 45.671 48.127 51.631 56.532 60.765 64.406 67.046 69.718 71.096 72.370 73.422
Yemen Rep. Asia 32.548 33.970 35.180 36.984 39.848 44.175 49.113 52.922 55.599 58.020 60.308 62.698
Zambia Africa 42.038 44.077 46.023 47.768 50.107 51.386 51.821 50.821 46.100 40.238 39.193 42.384
Zimbabwe Africa 48.451 50.469 52.358 53.995 55.635 57.674 60.363 62.351 60.377 46.809 39.989 43.487
[142 rows x 12 columns]
Notice here that we tell pivot() which columns to pull the names we wish our new columns to be named from the year variable,
and the values to populate those columns from the lifeExp variable.
We see that the resulting table has new columns by year, and the values populate it with country and continent dictating the rows.
The pandas melt() method allows us to “melt” a table from wide format to long format.
The code below converts our wide table back to the long format.
(
gapminder
.filter(['country', 'continent', 'year', 'lifeExp'])
.pivot(columns='year',
index=['country', 'continent'],
values='lifeExp')
.reset_index()
.melt(id_vars=['country', 'continent'],
value_name='lifeExp')
)
country continent year lifeExp
0 Afghanistan Asia 1952 28.801
1 Albania Europe 1952 55.230
2 Algeria Africa 1952 43.077
3 Angola Africa 1952 30.015
4 Argentina Americas 1952 62.485
... ... ... ... ...
1699 Vietnam Asia 2007 74.249
1700 West Bank and Gaza Asia 2007 73.422
1701 Yemen Rep. Asia 2007 62.698
1702 Zambia Africa 2007 42.384
1703 Zimbabwe Africa 2007 43.487
[1704 rows x 4 columns]
Before we move on to more data cleaning, let’s create the final gapminder data frame we will be working with for the rest of the lesson!
Final Americas 2007 gapminder dataset
- Read in the
gapminder_data.csvfile.- Filter out the year 2007 and the continent “Americas”.
- Drop the
yearandcontinentcolumns from the data frame.- Save the new data frame into a variable called
gapminder_2007.Solution:
gapminder_2007 = ( gapminder .query("year == 2007 and continent == 'Americas'") .drop(columns=['year', 'continent']) )country pop lifeExp gdpPercap 59 Argentina 40301927.0 75.320 12779.379640 143 Bolivia 9119152.0 65.554 3822.137084 179 Brazil 190010647.0 72.390 9065.800825 251 Canada 33390141.0 80.653 36319.235010 287 Chile 16284741.0 78.553 13171.638850 311 Colombia 44227550.0 72.889 7006.580419 359 Costa Rica 4133884.0 78.782 9645.061420 395 Cuba 11416987.0 78.273 8948.102923 443 Dominican Republic 9319622.0 72.235 6025.374752 455 Ecuador 13755680.0 74.994 6873.262326 479 El Salvador 6939688.0 71.878 5728.353514 611 Guatemala 12572928.0 70.259 5186.050003 647 Haiti 8502814.0 60.916 1201.637154 659 Honduras 7483763.0 70.198 3548.330846 791 Jamaica 2780132.0 72.567 7320.880262 995 Mexico 108700891.0 76.195 11977.574960 1115 Nicaragua 5675356.0 72.899 2749.320965 1187 Panama 3242173.0 75.537 9809.185636 1199 Paraguay 6667147.0 71.752 4172.838464 1211 Peru 28674757.0 71.421 7408.905561 1259 Puerto Rico 3942491.0 78.746 19328.709010 1559 Trinidad and Tobago 1056608.0 69.819 18008.509240 1619 United States 301139947.0 78.242 42951.653090 1631 Uruguay 3447496.0 76.384 10611.462990 1643 Venezuela 26084662.0 73.747 11415.805690
Awesome! This is the data frame we will be using later on in this lesson.
Reviewing Git and GitHub
Now that we have our gapminder data prepared, let’s use what we learned about git and GitHub in the previous lesson to add, commit, and push our changes.
Open Terminal/Git Bash, if you do not have it open already. First we’ll need to navigate to our un-report directory.
Let’s start by printing our current working directory and listing the items in the directory, to see where we are.
pwd
ls
Now, we’ll navigate to the un-report directory.
cd ~/Desktop/un-report
ls
To start, let’s pull to make sure our local repository is up to date.
git status
git pull
Not let’s add and commit our changes.
git status
git add
git status "gapminder_data_analysis.ipynb"
git commit -m "Create data analysis file"
Finally, let’s check our commits and then push the commits to GitHub.
git status
git log --online
git push
git status
Cleaning up data
Researchers are often pulling data from several sources, and the process of making data compatible with one another and prepared for analysis can be a large undertaking. Luckily, there are many functions that allow us to do this with pandas. We’ve been working with the gapminder dataset, which contains population and GDP data by year. In this section, we practice cleaning and preparing a second dataset containing CO2 emissions data by country and year, sourced from the UN.
It’s always good to go into data cleaning with a clear goal in mind.
Here, we’d like to prepare the CO2 UN data to be compatible with our gapminder data so we can directly compare GDP to CO2 emissions.
To make this work, we’d like a data frame that contains a column with the country name, and columns for different ways of measuring CO2 emissions.
We will also want the data to be collected as close to 2007 as possible (the last year we have data for in gapminder).
Let’s start with reading the data using pandas’s read_csv() function.
pd.read_csv("./data/co2-un-data.csv")
T24 CO2 emission estimates Unnamed: 2 Unnamed: 3 Unnamed: 4 Unnamed: 5 \
0 Region/Country/Area NaN Year Series Value Footnotes
1 8 Albania 1975 Emissions (thousand metric tons of carbon diox... 4338.3340 NaN
2 8 Albania 1985 Emissions (thousand metric tons of carbon diox... 6929.9260 NaN
3 8 Albania 1995 Emissions (thousand metric tons of carbon diox... 1848.5490 NaN
4 8 Albania 2005 Emissions (thousand metric tons of carbon diox... 3825.1840 NaN
... ... ... ... ... ... ...
2128 716 Zimbabwe 2005 Emissions per capita (metric tons of carbon di... 0.7940 NaN
2129 716 Zimbabwe 2010 Emissions per capita (metric tons of carbon di... 0.6720 NaN
2130 716 Zimbabwe 2015 Emissions per capita (metric tons of carbon di... 0.7490 NaN
2131 716 Zimbabwe 2016 Emissions per capita (metric tons of carbon di... 0.6420 NaN
2132 716 Zimbabwe 2017 Emissions per capita (metric tons of carbon di... 0.5880 NaN
Unnamed: 6
0 Source
1 International Energy Agency, IEA World Energy ...
2 International Energy Agency, IEA World Energy ...
3 International Energy Agency, IEA World Energy ...
4 International Energy Agency, IEA World Energy ...
... ...
2128 International Energy Agency, IEA World Energy ...
2129 International Energy Agency, IEA World Energy ...
2130 International Energy Agency, IEA World Energy ...
2131 International Energy Agency, IEA World Energy ...
2132 International Energy Agency, IEA World Energy ...
[2133 rows x 7 columns]
Looking at the table that is outputted above we can see that there appear to be two rows at the top of the file that contain information about the data in the table.
The first is a header that tells us the table number and its name.
Ideally, we’d skip that. We can do this using the skiprows argument in read_csv() by giving it a number of rows to skip.
pd.read_csv("./data/co2-un-data.csv", skiprows=1)
Region/Country/Area Unnamed: 1 Year Series Value Footnotes \
0 8 Albania 1975 Emissions (thousand metric tons of carbon diox... 4338.334 NaN
1 8 Albania 1985 Emissions (thousand metric tons of carbon diox... 6929.926 NaN
2 8 Albania 1995 Emissions (thousand metric tons of carbon diox... 1848.549 NaN
3 8 Albania 2005 Emissions (thousand metric tons of carbon diox... 3825.184 NaN
4 8 Albania 2010 Emissions (thousand metric tons of carbon diox... 3930.295 NaN
... ... ... ... ... ... ...
2127 716 Zimbabwe 2005 Emissions per capita (metric tons of carbon di... 0.794 NaN
2128 716 Zimbabwe 2010 Emissions per capita (metric tons of carbon di... 0.672 NaN
2129 716 Zimbabwe 2015 Emissions per capita (metric tons of carbon di... 0.749 NaN
2130 716 Zimbabwe 2016 Emissions per capita (metric tons of carbon di... 0.642 NaN
2131 716 Zimbabwe 2017 Emissions per capita (metric tons of carbon di... 0.588 NaN
Source
0 International Energy Agency, IEA World Energy ...
1 International Energy Agency, IEA World Energy ...
2 International Energy Agency, IEA World Energy ...
3 International Energy Agency, IEA World Energy ...
4 International Energy Agency, IEA World Energy ...
... ...
2127 International Energy Agency, IEA World Energy ...
2128 International Energy Agency, IEA World Energy ...
2129 International Energy Agency, IEA World Energy ...
2130 International Energy Agency, IEA World Energy ...
2131 International Energy Agency, IEA World Energy ...
[2132 rows x 7 columns]
Now the output table looks better.
Another thing we can do is to tell the read_csv() function what the column names should be with the names argument where we give it the column names we want as a Python list.
If we do this, then we need to skip 2 rows including the original column headings.
Let’s also save this data frame to co2_emissions_dirty so that we don’t have to read it every time we want to clean it even more.
co2_emissions_dirty = (
pd.read_csv("./data/co2-un-data.csv", skiprows=2,
names=['region', 'country', 'year', 'series', 'value', 'footnotes', 'source'])
)
co2_emissions_dirty
region country year series value footnotes \
0 8 Albania 1975 Emissions (thousand metric tons of carbon diox... 4338.334 NaN
1 8 Albania 1985 Emissions (thousand metric tons of carbon diox... 6929.926 NaN
2 8 Albania 1995 Emissions (thousand metric tons of carbon diox... 1848.549 NaN
3 8 Albania 2005 Emissions (thousand metric tons of carbon diox... 3825.184 NaN
4 8 Albania 2010 Emissions (thousand metric tons of carbon diox... 3930.295 NaN
... ... ... ... ... ... ...
2127 716 Zimbabwe 2005 Emissions per capita (metric tons of carbon di... 0.794 NaN
2128 716 Zimbabwe 2010 Emissions per capita (metric tons of carbon di... 0.672 NaN
2129 716 Zimbabwe 2015 Emissions per capita (metric tons of carbon di... 0.749 NaN
2130 716 Zimbabwe 2016 Emissions per capita (metric tons of carbon di... 0.642 NaN
2131 716 Zimbabwe 2017 Emissions per capita (metric tons of carbon di... 0.588 NaN
source
0 International Energy Agency, IEA World Energy ...
1 International Energy Agency, IEA World Energy ...
2 International Energy Agency, IEA World Energy ...
3 International Energy Agency, IEA World Energy ...
4 International Energy Agency, IEA World Energy ...
... ...
2127 International Energy Agency, IEA World Energy ...
2128 International Energy Agency, IEA World Energy ...
2129 International Energy Agency, IEA World Energy ...
2130 International Energy Agency, IEA World Energy ...
2131 International Energy Agency, IEA World Energy ...
[2132 rows x 7 columns]
Bonus: Another way to deal with the column names
Many data analysts prefer to have their column names be in all lower case. We can apply the
rename()method to set all of the column names to lower case.( pd.read_csv("./data/co2-un-data.csv", skiprows=1) .rename(columns=str.lower) )region/country/area unnamed: 1 year series value footnotes \ 0 8 Albania 1975 Emissions (thousand metric tons of carbon diox... 4338.334 NaN 1 8 Albania 1985 Emissions (thousand metric tons of carbon diox... 6929.926 NaN 2 8 Albania 1995 Emissions (thousand metric tons of carbon diox... 1848.549 NaN 3 8 Albania 2005 Emissions (thousand metric tons of carbon diox... 3825.184 NaN 4 8 Albania 2010 Emissions (thousand metric tons of carbon diox... 3930.295 NaN ... ... ... ... ... ... ... 2127 716 Zimbabwe 2005 Emissions per capita (metric tons of carbon di... 0.794 NaN 2128 716 Zimbabwe 2010 Emissions per capita (metric tons of carbon di... 0.672 NaN 2129 716 Zimbabwe 2015 Emissions per capita (metric tons of carbon di... 0.749 NaN 2130 716 Zimbabwe 2016 Emissions per capita (metric tons of carbon di... 0.642 NaN 2131 716 Zimbabwe 2017 Emissions per capita (metric tons of carbon di... 0.588 NaN source 0 International Energy Agency, IEA World Energy ... 1 International Energy Agency, IEA World Energy ... 2 International Energy Agency, IEA World Energy ... 3 International Energy Agency, IEA World Energy ... 4 International Energy Agency, IEA World Energy ... ... ... 2127 International Energy Agency, IEA World Energy ... 2128 International Energy Agency, IEA World Energy ... 2129 International Energy Agency, IEA World Energy ... 2130 International Energy Agency, IEA World Energy ... 2131 International Energy Agency, IEA World Energy ... [2132 rows x 7 columns]
We previously saw how we can subset columns from a data frame using the select function. There are a lot of columns with extraneous information in this dataset, let’s subset out the columns we are interested in.
Reviewing selecting columns
Select the country, year, series, and value columns from our dataset.
Solution:
( co2_emissions_dirty .filter(['country', 'year', 'series', 'value']) )country year series value 0 Albania 1975 Emissions (thousand metric tons of carbon diox... 4338.334 1 Albania 1985 Emissions (thousand metric tons of carbon diox... 6929.926 2 Albania 1995 Emissions (thousand metric tons of carbon diox... 1848.549 3 Albania 2005 Emissions (thousand metric tons of carbon diox... 3825.184 4 Albania 2010 Emissions (thousand metric tons of carbon diox... 3930.295 ... ... ... ... ... 2127 Zimbabwe 2005 Emissions per capita (metric tons of carbon di... 0.794 2128 Zimbabwe 2010 Emissions per capita (metric tons of carbon di... 0.672 2129 Zimbabwe 2015 Emissions per capita (metric tons of carbon di... 0.749 2130 Zimbabwe 2016 Emissions per capita (metric tons of carbon di... 0.642 2131 Zimbabwe 2017 Emissions per capita (metric tons of carbon di... 0.588 [2132 rows x 4 columns]
The series column has two methods of quantifying CO2 emissions - “Emissions (thousand metric tons of carbon dioxide)” and “Emissions per capita (metric tons of carbon dioxide)”.
Those are long titles that we’d like to shorten to make them easier to work with.
We can shorten them to “emissions_total” and “emissions_percap” using the recode function.
We can achieve this by applying the pandas replace() method to replace the values.
When using the replace() method we need to tell it which column we want to replace values with and then what is the old value (e.g. “Emissions (thousand metric tons of carbon dioxide)”) and new values (e.g. “emissions_total”).
(
co2_emissions_dirty
.filter(['country', 'year', 'series', 'value'])
.replace({'series': {"Emissions (thousand metric tons of carbon dioxide)":"emissions_total",
"Emissions per capita (metric tons of carbon dioxide)":"emissions_percap"},
})
)
country year series value
0 Albania 1975 emissions_total 4338.334
1 Albania 1985 emissions_total 6929.926
2 Albania 1995 emissions_total 1848.549
3 Albania 2005 emissions_total 3825.184
4 Albania 2010 emissions_total 3930.295
... ... ... ... ...
2127 Zimbabwe 2005 emissions_percap 0.794
2128 Zimbabwe 2010 emissions_percap 0.672
2129 Zimbabwe 2015 emissions_percap 0.749
2130 Zimbabwe 2016 emissions_percap 0.642
2131 Zimbabwe 2017 emissions_percap 0.588
[2132 rows x 4 columns]
Recall that we’d like to have separate columns for the two ways that we CO2 emissions data.
To achieve this, we’ll apply the pivot method that we used previously.
The columns we want to spread out are “series” (i.e. the columns argument) and “value” (i.e. the value argument).
(
co2_emissions_dirty
.filter(['country', 'year', 'series', 'value'])
.replace({'series': {"Emissions (thousand metric tons of carbon dioxide)":"emissions_total",
"Emissions per capita (metric tons of carbon dioxide)":"emissions_percap"},
})
.pivot(index=['country', 'year'], columns='series', values='value')
.reset_index()
)
series country year emissions_percap emissions_total
0 Albania 1975 1.804 4338.334
1 Albania 1985 2.337 6929.926
2 Albania 1995 0.580 1848.549
3 Albania 2005 1.270 3825.184
4 Albania 2010 1.349 3930.295
... ... ... ... ...
1061 Zimbabwe 2005 0.794 10272.774
1062 Zimbabwe 2010 0.672 9464.714
1063 Zimbabwe 2015 0.749 11822.362
1064 Zimbabwe 2016 0.642 10368.900
1065 Zimbabwe 2017 0.588 9714.938
[1066 rows x 4 columns]
Excellent! The last step before we can join this data frame is to get the most data that is for the year closest to 2007 so we can make a more direct comparison to the most recent data we have from gapminder. For the sake of time, we’ll just tell you that we want data from 2005.
Bonus: How did we determine that 2005 is the closest year to 2007?
We want to make sure we pick a year that is close to 2005, but also a year that has a decent amount of data to work with. One useful tool is the
value_counts()method, which will tell us how many times a value is repeated in a column of a data frame. Let’s use this function on the year column to see which years we have data for and to tell us whether we have a good number of countries represented in that year.( co2_emissions_dirty .filter(['country', 'year', 'series', 'value']) .replace({'series': {"Emissions (thousand metric tons of carbon dioxide)> ":"emissions_total", "Emissions per capita (metric tons of carbon dioxide)> ":"emissions_percap"}, }) .pivot(index=['country', 'year'], columns='series', values='value') .reset_index() .value_counts(['year']) .sort_index() )year 1975 111 1985 113 1995 136 2005 140 2010 140 2015 142 2016 142 2017 142 Name: count, dtype: int64It looks like we have data for 140 countries in 2005 and 2010. We chose 2005 because it is closer to 2007.
Filtering rows and removing columns
Filter out data from 2005 and then drop the year column. (Since we will have only data from one year, it is now irrelevant.)
Solution:
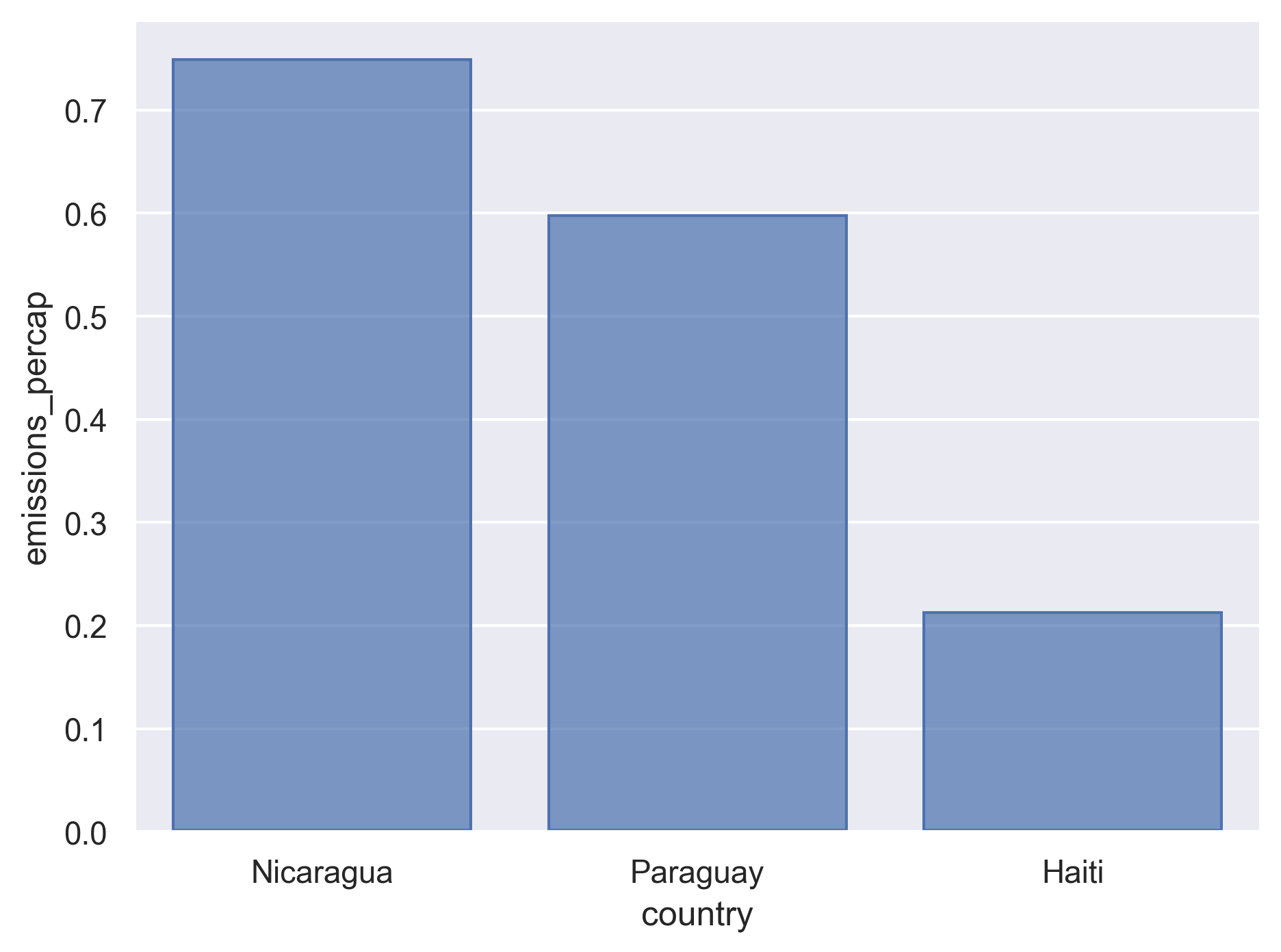
( co2_emissions_dirty .filter(['country', 'year', 'series', 'value']) .replace({'series': {"Emissions (thousand metric tons of carbon dioxide)> > ":"emissions_total", "Emissions per capita (metric tons of carbon dioxide)> > ":"emissions_percap"}, }) .pivot(index=['country', 'year'], columns='series', values='value') .reset_index() .query("year == 2005") .drop(columns='year') )series country emissions_percap emissions_total 3 Albania 1.270 3825.184 11 Algeria 2.327 77474.130 19 Angola 0.314 6146.691 27 Argentina 3.819 149476.040 33 Armenia 1.385 4129.845 ... ... ... ... 1029 Venezuela (Boliv. Rep. of) 5.141 137701.548 1037 Viet Nam 0.940 79230.185 1045 Yemen 0.915 18836.222 1053 Zambia 0.176 2120.692 1061 Zimbabwe 0.794 10272.774 [140 rows x 3 columns]
Finally, let’s go ahead and assign the output of this code chunk, which is the cleaned data frame, to a variable name:
co2_emissions = (
co2_emissions_dirty
.filter(['country', 'year', 'series', 'value'])
.replace({'series': {'Emissions (thousand metric tons of carbon dioxide)':'emissions_total',
'Emissions per capita (metric tons of carbon dioxide)':'emissions_percap'},
})
.pivot(index=['country', 'year'], columns='series', values='value')
.reset_index()
.query("year == 2005")
.drop(columns='year')
)
Joining data frames
Now we’re ready to join our CO2 emissions data to the gapminder data. Previously we saw that we could read in and query the gapminder data like this to get the data from the Americas for 2007 so we can create a new data frame with our filtered data:
gapminder_2007 = (
gapminder
.query("year == 2007 and continent == 'Americas'")
.drop(columns=['year', 'continent'])
)
Look at the data in co2_emissions and gapminder_data_2007.
If you had to merge these two data frames together, which column would you use to merge them together?
If you said “country” - good job!
We’ll call country our “key”. Now, when we join them together, can you think of any problems we might run into when we merge things? We might not have CO2 emissions data for all of the countries in the gapminder dataset and vice versa. Also, a country might be represented in both data frames but not by the same name in both places. As an example, write down the name of the country that the University of Michigan is in - we’ll come back to your answer shortly!
pandas has a number of tools for joining data frames together depending on what we want to do with the rows of the data of countries that are not represented in both data frames. Here we’ll be using “inner join” and “outer join”.
In an “inner join”, the new data frame only has those rows where the same key is found in both data frames. This is a very commonly used join.

Bonus: Other pandas join methods
There are other types of join too. For a left join, if the key is present in the left hand data frame, it will appear in the output, even if it is not found in the the right hand data frame. For a right join, the opposite is true. For a outer (or full) join, all possible keys are included in the output data frame.
Let’s give the merge() method a try.
(
gapminder_2007
.merge(co2_emissions, how='inner', on='country')
)
country pop lifeExp gdpPercap emissions_percap emissions_total
0 Argentina 40301927.0 75.320 12779.379640 3.819 149476.040
1 Brazil 190010647.0 72.390 9065.800825 1.667 311623.799
2 Canada 33390141.0 80.653 36319.235010 16.762 540431.495
3 Chile 16284741.0 78.553 13171.638850 3.343 54434.634
4 Colombia 44227550.0 72.889 7006.580419 1.238 53585.300
5 Costa Rica 4133884.0 78.782 9645.061420 1.286 5463.059
6 Cuba 11416987.0 78.273 8948.102923 2.220 25051.431
7 Dominican Republic 9319622.0 72.235 6025.374752 1.897 17522.139
8 Ecuador 13755680.0 74.994 6873.262326 1.742 23926.725
9 El Salvador 6939688.0 71.878 5728.353514 1.037 6252.815
10 Guatemala 12572928.0 70.259 5186.050003 0.811 10621.597
11 Haiti 8502814.0 60.916 1201.637154 0.214 1980.992
12 Honduras 7483763.0 70.198 3548.330846 0.976 7192.737
13 Jamaica 2780132.0 72.567 7320.880262 3.746 10281.648
14 Mexico 108700891.0 76.195 11977.574960 3.854 412385.135
15 Nicaragua 5675356.0 72.899 2749.320965 0.750 4032.083
16 Panama 3242173.0 75.537 9809.185636 2.035 6776.118
17 Paraguay 6667147.0 71.752 4172.838464 0.599 3472.665
18 Peru 28674757.0 71.421 7408.905561 1.037 28632.888
19 Trinidad and Tobago 1056608.0 69.819 18008.509240 13.243 17175.823
20 Uruguay 3447496.0 76.384 10611.462990 1.549 5151.871
Do you see that we now have data from both data frames joined together?
One thing to notice is that gapminder data had 25 rows, but the output of our join only had 21. Let’s investigate. It appears that there must have been countries in the gapminder data that did not appear in our CO2 emission data.
Let’s do another merge for this, this time with an outer join.
If we set the indicator argument to True, it will add a new column called _merge to the merged data, and the value indicates whether a particular record appeared at left_only, right_only, or both.
Then we can do a query to show the data for the keys on the left that are missing from the data frame on the right.
(
gapminder_2007
.merge(co2_emissions, how='outer', on='country', indicator=True)
.query("_merge == 'left_only'")
)
country pop lifeExp gdpPercap emissions_percap emissions_total _merge
1 Bolivia 9119152.0 65.554 3822.137084 NaN NaN left_only
20 Puerto Rico 3942491.0 78.746 19328.709010 NaN NaN left_only
22 United States 301139947.0 78.242 42951.653090 NaN NaN left_only
24 Venezuela 26084662.0 73.747 11415.805690 NaN NaN left_only
We can see that the CO2 emission data were missing for Bolivia, Puerto Rico, United States, and Venezuela.
We can query the CO2 emission data to check if there are records containing these names.
Note we can split a long string by adding a backslash \ (it’s called a line continuation character) at the end of each line.
The string will continue on the next line as if it were a single line.
(
co2_emissions
.query("country.str.contains('Bolivia') or \
country.str.contains('Puerto Rico') or \
country.str.contains('United States') or \
country.str.contains('Venezuela')")
)
series country emissions_percap emissions_total
101 Bolivia (Plurin. State of) 0.984 8975.809
1007 United States of America 19.268 5703220.175
1029 Venezuela (Boliv. Rep. of) 5.141 137701.548
From the outputs above we can see that Bolivia, the United States, and Venezuela are called different things in the CO2 emission data.
Puerto Rico isn’t a country; it’s part of the United States.
We can apply the replace() method to these country names in the CO2 emission data so that the country names for Bolivia, United States, and Venezuela, match those in the gapminder data.
(
co2_emissions
.replace({'country':{'Bolivia (Plurin. State of)':'Bolivia',
'United States of America':'United States',
'Venezuela (Boliv. Rep. of)':'Venezuela'}
})
)
series country emissions_percap emissions_total
3 Albania 1.270 3825.184
11 Algeria 2.327 77474.130
19 Angola 0.314 6146.691
27 Argentina 3.819 149476.040
33 Armenia 1.385 4129.845
... ... ... ...
1029 Venezuela 5.141 137701.548
1037 Viet Nam 0.940 79230.185
1045 Yemen 0.915 18836.222
1053 Zambia 0.176 2120.692
1061 Zimbabwe 0.794 10272.774
[140 rows x 3 columns]
(
gapminder_2007
.merge(co2_emissions.replace({'country':{'Bolivia (Plurin. State of)':'Bolivia',
'United States of America':'United States',
'Venezuela (Boliv. Rep. of)':'Venezuela'}
}),
how='outer', on='country', indicator=True)
.query("_merge == 'left_only'")
)
country pop lifeExp gdpPercap emissions_percap emissions_total _merge
20 Puerto Rico 3942491.0 78.746 19328.70901 NaN NaN left_only
Now we see that the replacement of the country names enabled the join for all countries in the gapminder, and we are left with Puerto Rico.
In the next exercise, let’s replace the name Puerto Rico to the United States in the gapminder data and then use the groupby() method to aggregate the data.
We’ll use the population data to weight the life expectancy and GDP values.
In the gapminder data, let’s first replace the name Puerto Rico to the United States.
(
gapminder_2007
.replace({'country':{'Puerto Rico':'United States'}})
)
country pop lifeExp gdpPercap
59 Argentina 40301927.0 75.320 12779.379640
143 Bolivia 9119152.0 65.554 3822.137084
179 Brazil 190010647.0 72.390 9065.800825
251 Canada 33390141.0 80.653 36319.235010
287 Chile 16284741.0 78.553 13171.638850
311 Colombia 44227550.0 72.889 7006.580419
359 Costa Rica 4133884.0 78.782 9645.061420
395 Cuba 11416987.0 78.273 8948.102923
443 Dominican Republic 9319622.0 72.235 6025.374752
455 Ecuador 13755680.0 74.994 6873.262326
479 El Salvador 6939688.0 71.878 5728.353514
611 Guatemala 12572928.0 70.259 5186.050003
647 Haiti 8502814.0 60.916 1201.637154
659 Honduras 7483763.0 70.198 3548.330846
791 Jamaica 2780132.0 72.567 7320.880262
995 Mexico 108700891.0 76.195 11977.574960
1115 Nicaragua 5675356.0 72.899 2749.320965
1187 Panama 3242173.0 75.537 9809.185636
1199 Paraguay 6667147.0 71.752 4172.838464
1211 Peru 28674757.0 71.421 7408.905561
1259 United States 3942491.0 78.746 19328.709010
1559 Trinidad and Tobago 1056608.0 69.819 18008.509240
1619 United States 301139947.0 78.242 42951.653090
1631 Uruguay 3447496.0 76.384 10611.462990
1643 Venezuela 26084662.0 73.747 11415.805690
Now we have to group Puerto Rico and the US together, aggregating and calculating the data for all of the other columns. This is a little tricky - we will need a populated-weighted mean of lifeExp and gdpPercap.
(
gapminder_2007
.replace({'country':{'Puerto Rico':'United States'}})
.groupby('country')
.apply(lambda df: pd.Series({'pop': np.sum(df['pop']),
'gdpPercap': np.sum(df['gdpPercap'] * df['pop']) / np.sum(df['pop']),
'lifeExp': np.sum(df['lifeExp'] * df['pop']) / np.sum(df['pop']),
}))
)
pop gdpPercap lifeExp
country
Argentina 40301927.0 12779.379640 75.320000
Bolivia 9119152.0 3822.137084 65.554000
Brazil 190010647.0 9065.800825 72.390000
Canada 33390141.0 36319.235010 80.653000
Chile 16284741.0 13171.638850 78.553000
Colombia 44227550.0 7006.580419 72.889000
Costa Rica 4133884.0 9645.061420 78.782000
Cuba 11416987.0 8948.102923 78.273000
Dominican Republic 9319622.0 6025.374752 72.235000
Ecuador 13755680.0 6873.262326 74.994000
El Salvador 6939688.0 5728.353514 71.878000
Guatemala 12572928.0 5186.050003 70.259000
Haiti 8502814.0 1201.637154 60.916000
Honduras 7483763.0 3548.330846 70.198000
Jamaica 2780132.0 7320.880262 72.567000
Mexico 108700891.0 11977.574960 76.195000
Nicaragua 5675356.0 2749.320965 72.899000
Panama 3242173.0 9809.185636 75.537000
Paraguay 6667147.0 4172.838464 71.752000
Peru 28674757.0 7408.905561 71.421000
Trinidad and Tobago 1056608.0 18008.509240 69.819000
United States 305082438.0 42646.380702 78.248513
Uruguay 3447496.0 10611.462990 76.384000
Venezuela 26084662.0 11415.805690 73.747000
Let’s check to see if it worked!
(
gapminder_2007
.replace({'country':{'Puerto Rico': 'United States'}})
.groupby('country')
.apply(lambda df: pd.Series({'pop': np.sum(df['pop']),
'gdpPercap': np.sum(df['gdpPercap'] * df['pop']) / np.sum(df['pop']),
'lifeExp': np.sum(df['lifeExp'] * df['pop']) / np.sum(df['pop']),
}))
.merge(co2_emissions.replace({'country': {"Bolivia (Plurin. State of)":"Bolivia",
"United States of America":"United States",
"Venezuela (Boliv. Rep. of)":"Venezuela"}}),
how='outer', on='country', indicator=True)
.query("_merge == 'left_only'")
)
Empty DataFrame
Columns: [country, pop, gdpPercap, lifeExp, emissions_percap, emissions_total, _merge]
Index: []
Now the output above returns an empty data frame, which tells us that we have reconciled all of the keys from the gapminder data with the data in the CO2 emission data.
Finally, let’s merge the data with inner join to create a new data frame.
gapminder_co2 = (
gapminder_2007
.replace({'country':{'Puerto Rico': 'United States'}})
.groupby('country')
.apply(lambda df: pd.Series({'pop': np.sum(df['pop']),
'gdpPercap': np.sum(df['gdpPercap'] * df['pop']) / np.sum(df['pop']),
'lifeExp': np.sum(df['lifeExp'] * df['pop']) / np.sum(df['pop']),
}))
.merge(co2_emissions.replace({'country': {"Bolivia (Plurin. State of)":"Bolivia",
"United States of America":"United States",
"Venezuela (Boliv. Rep. of)":"Venezuela"}}),
how='inner', on='country')
)
One last thing! What if we’re interested in distinguishing between countries in North America and South America? We want to create two groups - Canada, the United States, and Mexico in one and the other countries in another.
We can apply the assign() method to add a new column and use the numpy function np.where() to help us define the region.
(
gapminder_co2
.assign(region=lambda df: np.where(df['country'].isin(['Canada', 'United States', 'Mexico']), 'north', 'south'))
)
country pop gdpPercap lifeExp emissions_percap emissions_total region
0 Argentina 40301927.0 12779.379640 75.320000 3.819 149476.040 south
1 Bolivia 9119152.0 3822.137084 65.554000 0.984 8975.809 south
2 Brazil 190010647.0 9065.800825 72.390000 1.667 311623.799 south
3 Canada 33390141.0 36319.235010 80.653000 16.762 540431.495 north
4 Chile 16284741.0 13171.638850 78.553000 3.343 54434.634 south
5 Colombia 44227550.0 7006.580419 72.889000 1.238 53585.300 south
6 Costa Rica 4133884.0 9645.061420 78.782000 1.286 5463.059 south
7 Cuba 11416987.0 8948.102923 78.273000 2.220 25051.431 south
8 Dominican Republic 9319622.0 6025.374752 72.235000 1.897 17522.139 south
9 Ecuador 13755680.0 6873.262326 74.994000 1.742 23926.725 south
10 El Salvador 6939688.0 5728.353514 71.878000 1.037 6252.815 south
11 Guatemala 12572928.0 5186.050003 70.259000 0.811 10621.597 south
12 Haiti 8502814.0 1201.637154 60.916000 0.214 1980.992 south
13 Honduras 7483763.0 3548.330846 70.198000 0.976 7192.737 south
14 Jamaica 2780132.0 7320.880262 72.567000 3.746 10281.648 south
15 Mexico 108700891.0 11977.574960 76.195000 3.854 412385.135 north
16 Nicaragua 5675356.0 2749.320965 72.899000 0.750 4032.083 south
17 Panama 3242173.0 9809.185636 75.537000 2.035 6776.118 south
18 Paraguay 6667147.0 4172.838464 71.752000 0.599 3472.665 south
19 Peru 28674757.0 7408.905561 71.421000 1.037 28632.888 south
20 Trinidad and Tobago 1056608.0 18008.509240 69.819000 13.243 17175.823 south
21 United States 305082438.0 42646.380702 78.248513 19.268 5703220.175 north
22 Uruguay 3447496.0 10611.462990 76.384000 1.549 5151.871 south
23 Venezuela 26084662.0 11415.805690 73.747000 5.141 137701.548 south
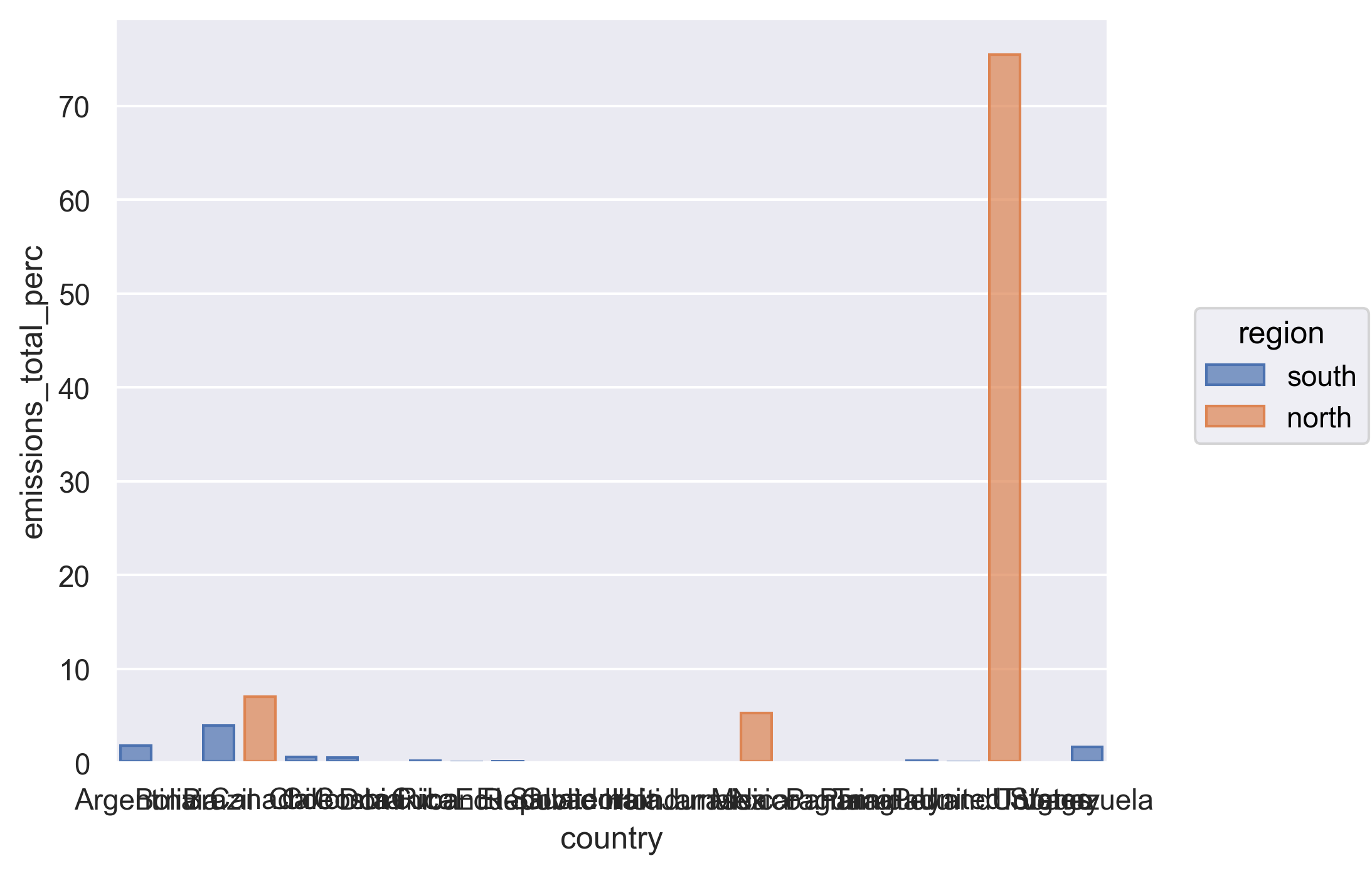
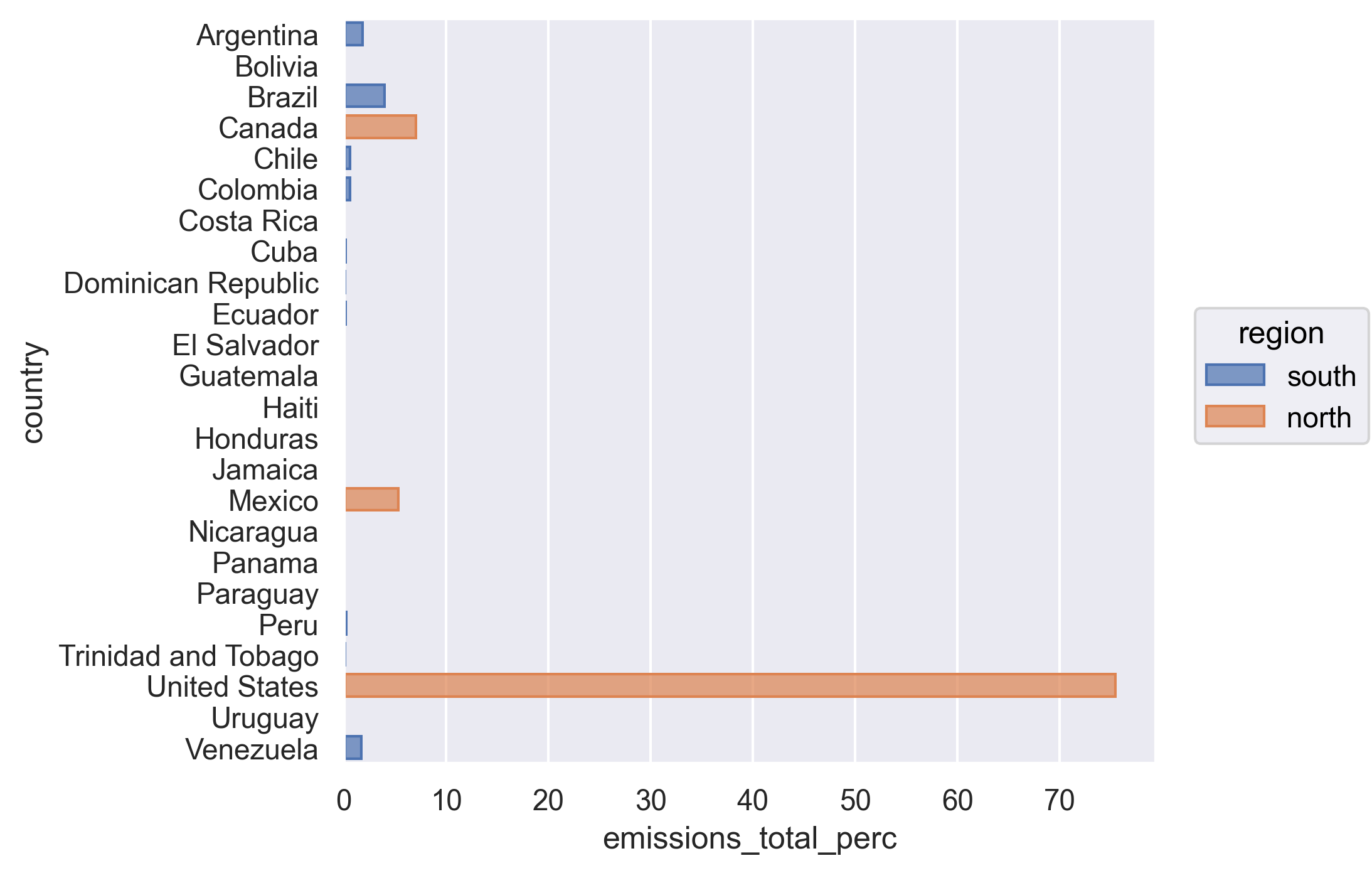
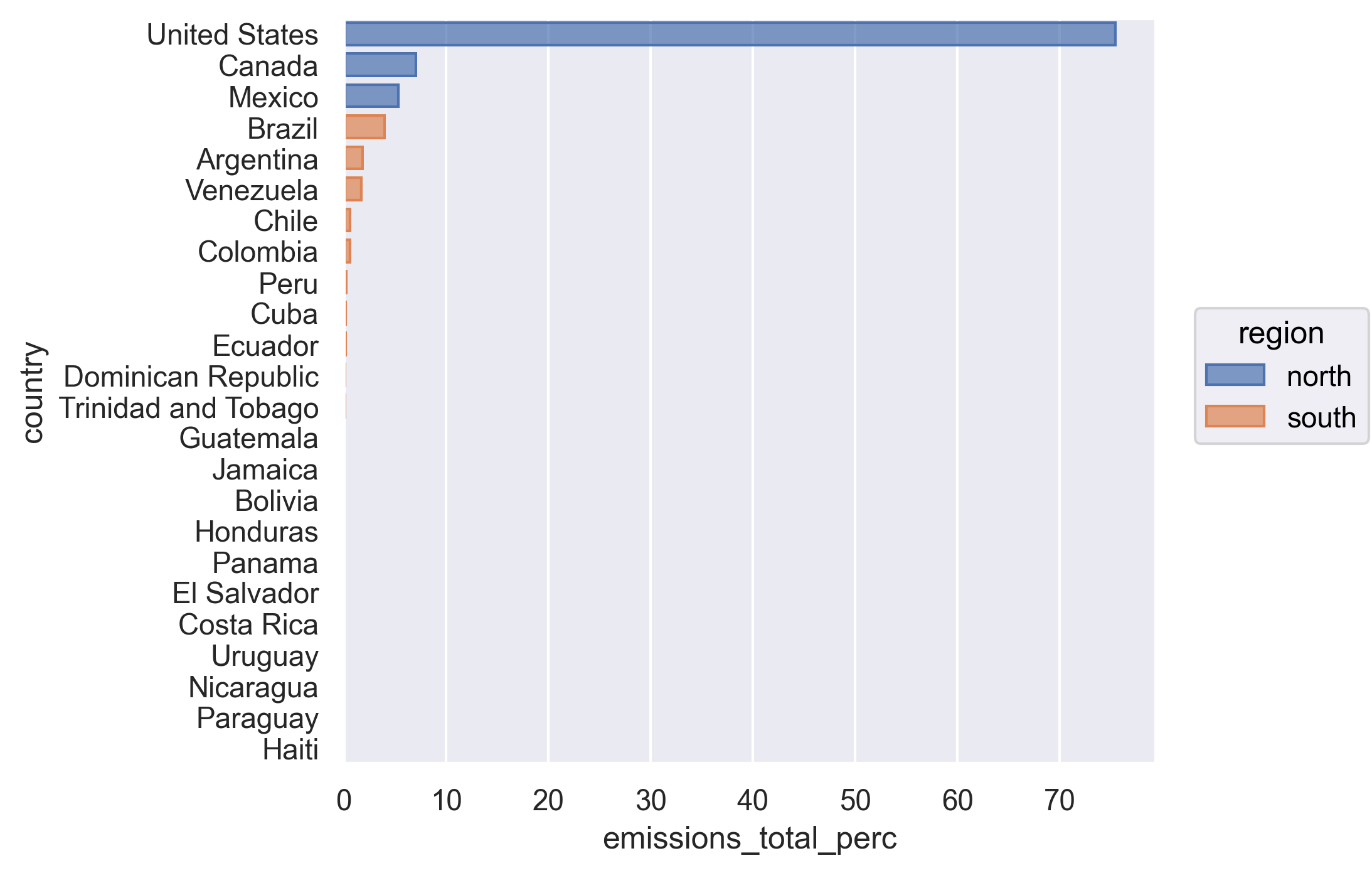
Let’s look at the output - see how the Canada, US, and Mexico rows are all labeled as “north” and everything else is labeled as “south”.
We have reached our data cleaning goals!
One of the best aspects of doing all of these steps coded in Python is that our efforts are reproducible, and the raw data is maintained.
With good documentation of data cleaning and analysis steps, we could easily share our work with another researcher who would be able to repeat what we’ve done.
However, it’s also nice to have a saved csv copy of our clean data.
That way we can access it later without needing to redo our data cleaning,
and we can also share the cleaned data with collaborators.
We can apply the to_csv method to a data frame to save it to a CSV file.
(
gapminder_co2
.assign(region=lambda df: np.where(df['country'].isin(['Canada', 'United States', 'Mexico']), 'north', 'south'))
.to_csv("./data/gapminder_co2.csv")
)
Great - Now we can move on to the analysis!
Analyzing combined data
For our analysis, we have two questions we’d like to answer. First, is there a relationship between the GDP of a country and the amount of CO2 emitted (per capita)? Second, Canada, the United States, and Mexico account for nearly half of the population of the Americas. What percent of the total CO2 production do they account for?
To answer the first question, we’ll plot the CO2 emitted (on a per capita basis) against the GDP (on a per capita basis) using a scatter plot:
import seaborn.objects as so
(
so.Plot(gapminder_co2, x='gdpPercap', y='emissions_percap')
.add(so.Dot())
.label(x="GDP (per capita)",
y="CO2 emitted (per capita)",
title="There is a strong association between a nation's GDP \nand the amount of CO2 it produces")
)
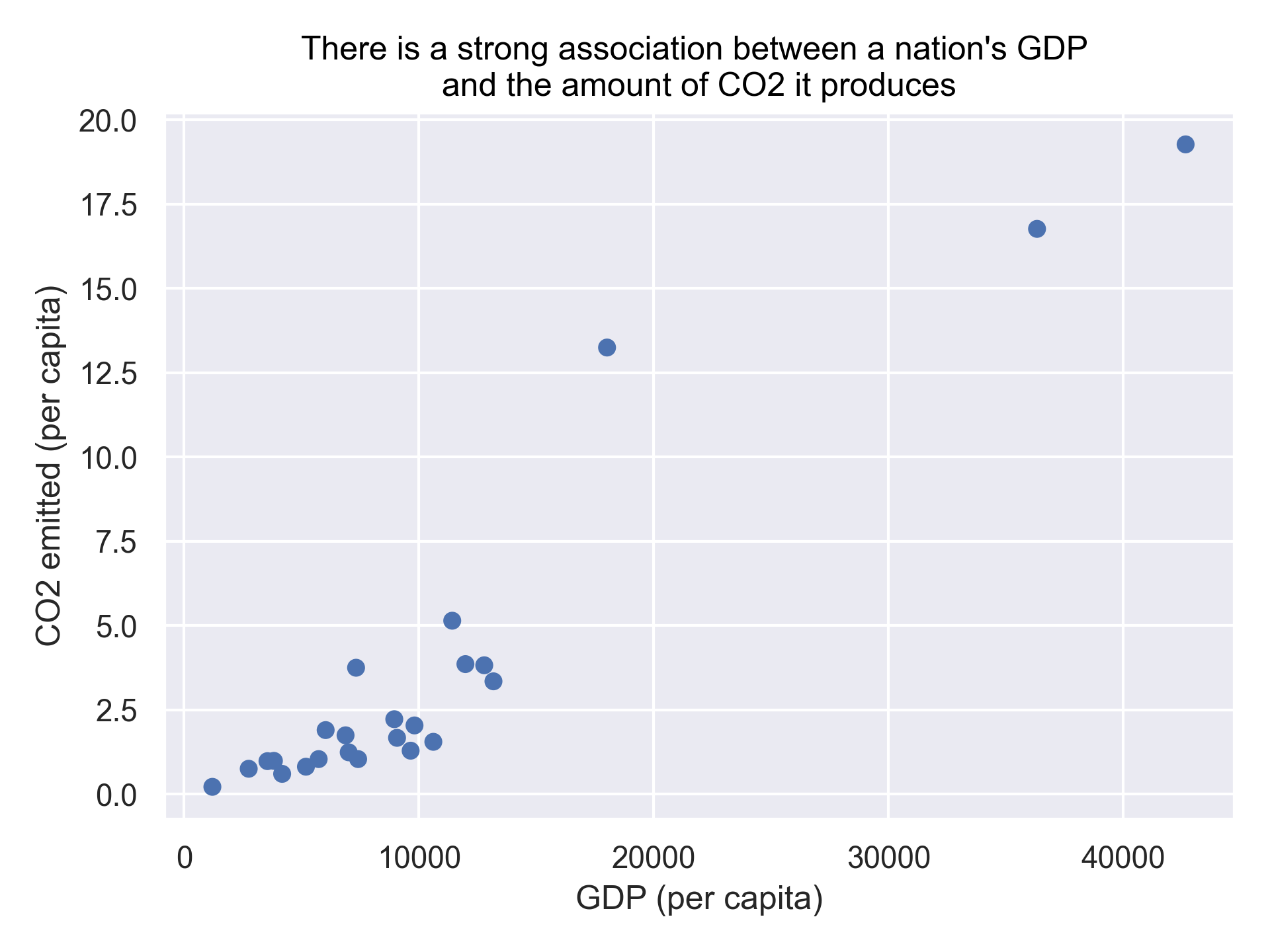
Tip: Notice we used the \n in our title to get a new line to prevent it from getting cut off.
To help clarify the association, we can add a fitted line representing a 3rd order polynomial regression model.
(
so.Plot(gapminder_co2, x='gdpPercap', y='emissions_percap')
.add(so.Dot(), label='data')
.add(so.Line(color='red'), so.PolyFit(order=3), label='model')
.label(x="GDP (per capita)",
y="CO2 emitted (per capita)",
title="There is a strong association between a nation's GDP \nand the amount of CO2 it produces")
)
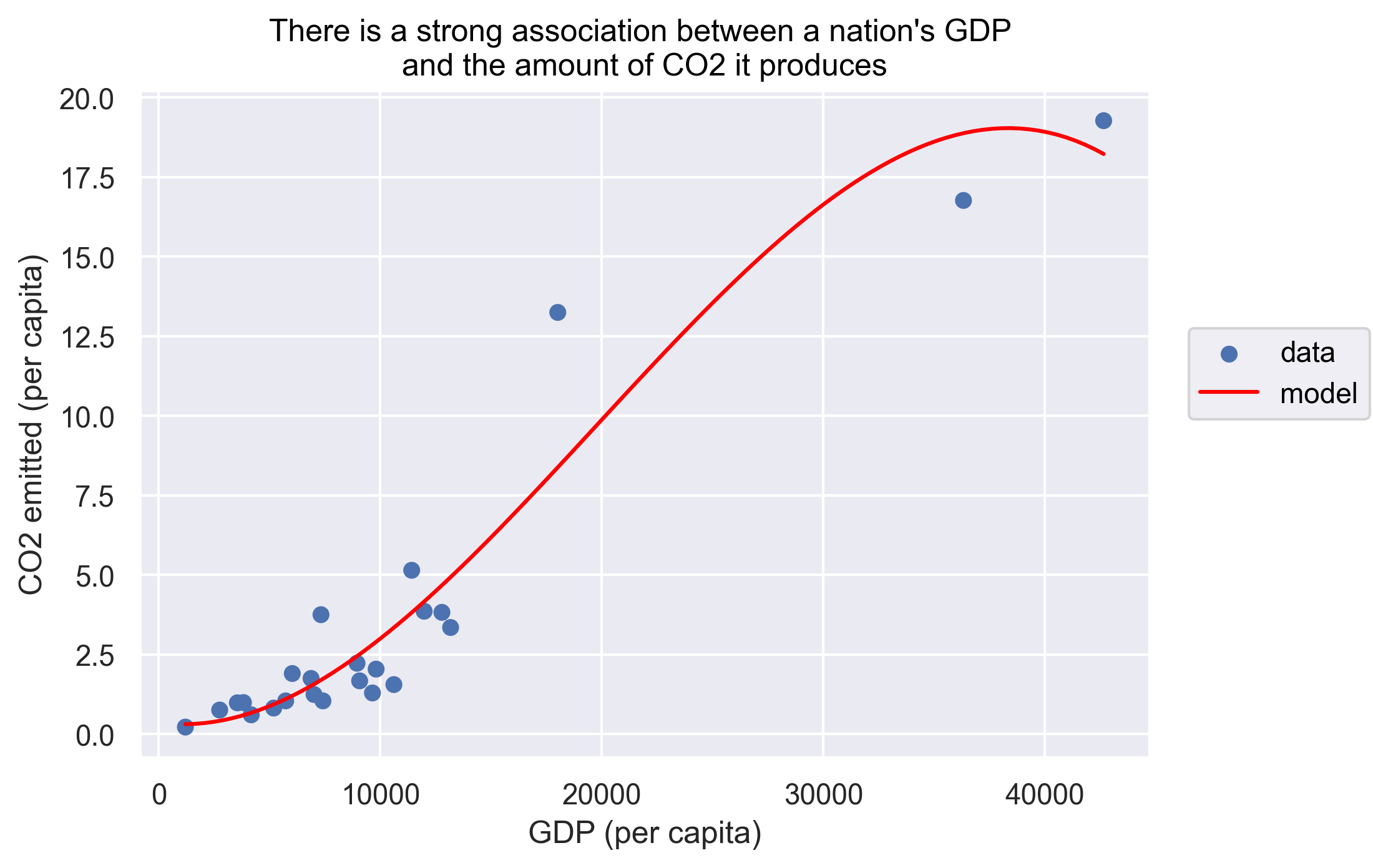
We can force the line to be straight using order=1 as an argument to so.PolyFit.
(
so.Plot(gapminder_co2, x='gdpPercap', y='emissions_percap')
.add(so.Dot(), label='data')
.add(so.Line(color='red'), so.PolyFit(order=1), label='model')
.label(x="GDP (per capita)",
y="CO2 emitted (per capita)",
title="There is a strong association between a nation's GDP \nand the amount of CO2 it produces")
)
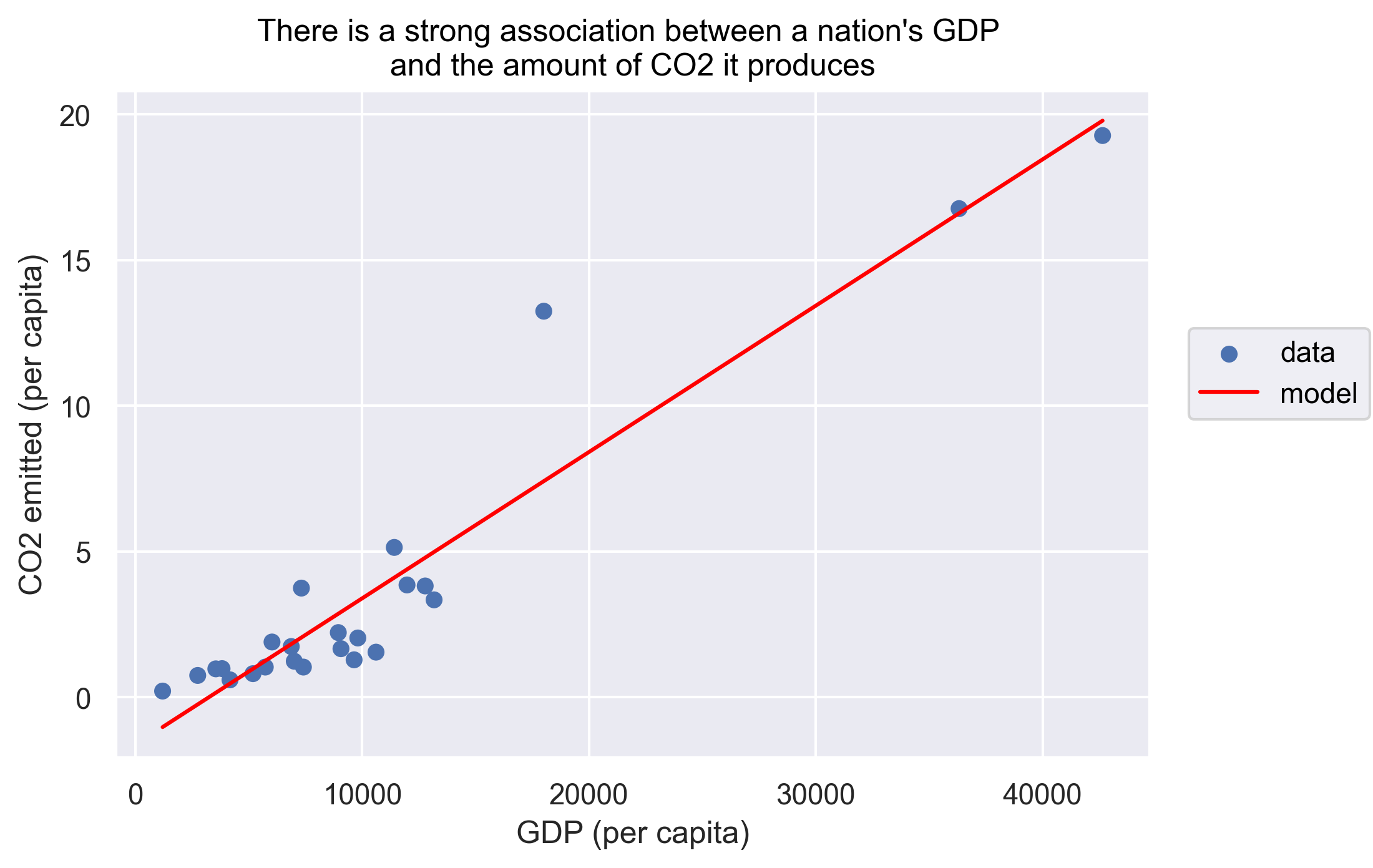
In addition, we see that only two or three countries have very high GDP/emissions, while the rest of the countries are cluttered in the lower ranges of the axes. To make it easier to see the relationship we can set the x and y axis to a logarithmic scale. Lastly, we can also add a text layer that displays the country names next to the markers.
(
so.Plot(gapminder_co2, x='gdpPercap', y='emissions_percap', text='country')
.add(so.Dot(alpha=.8, pointsize=8))
.add(so.Text(color='gray', valign='bottom', fontsize=10))
.scale(x='log', y='log')
.label(x="GDP (per capita)",
y="CO2 emitted (per capita)",
title="There is a strong association between a nation's GDP \nand the amount of CO2 it produces")
.limit(x=(None, 70_000), y=(None, 30))
)
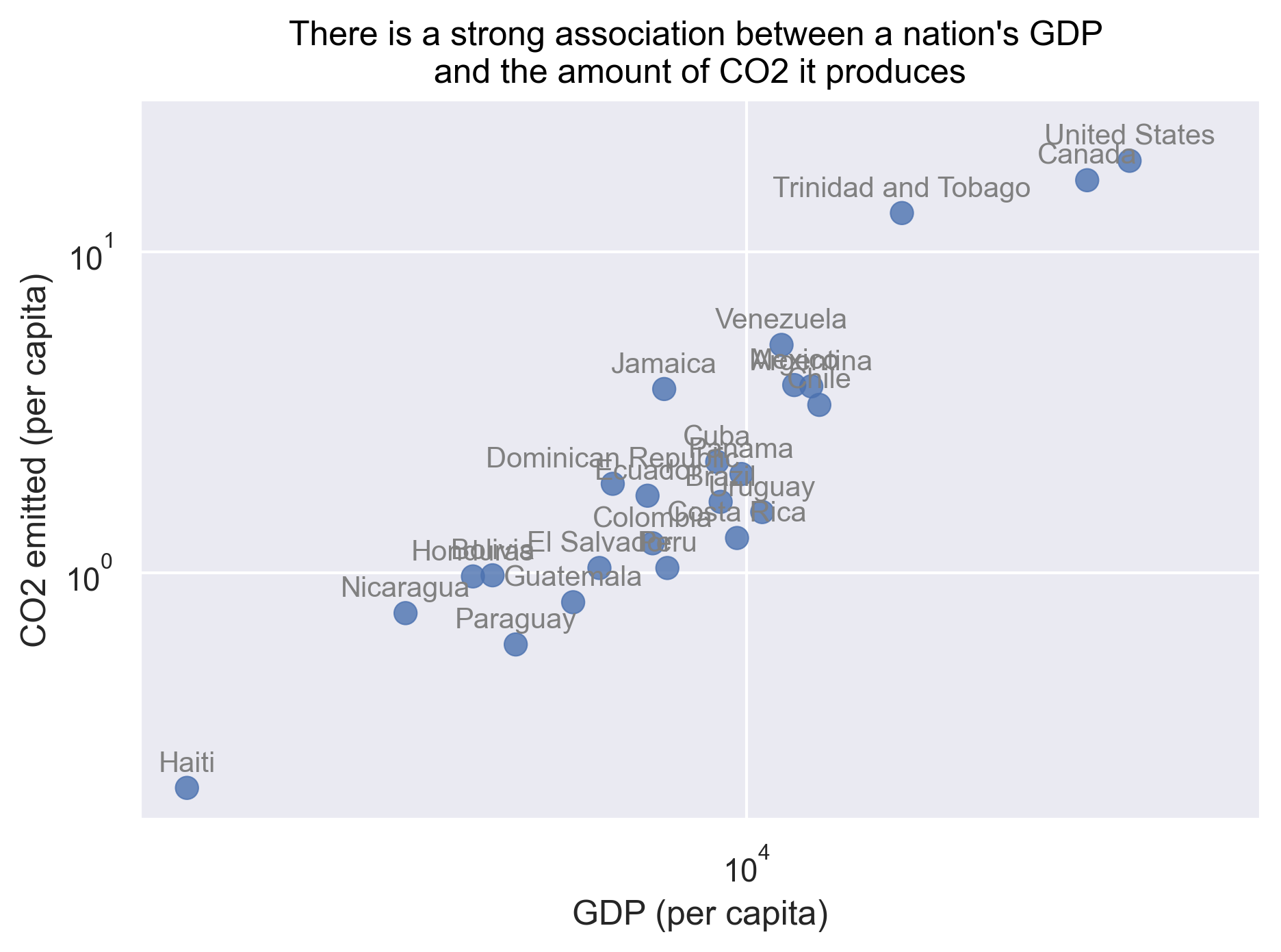
To answer our first question, as the title of our plot indicates there is indeed a strong association between a nation’s GDP and the amount of CO2 it produces.
For the second question, we want to create two groups - Canada, the United States, and Mexico in one and the other countries in another.
(
gapminder_co2
.assign(region=lambda df: np.where(df['country'].isin(['Canada', 'United States', 'Mexico']), 'north', 'south'))
)
country pop gdpPercap lifeExp emissions_percap emissions_total region
0 Argentina 40301927.0 12779.379640 75.320000 3.819 149476.040 south
1 Bolivia 9119152.0 3822.137084 65.554000 0.984 8975.809 south
2 Brazil 190010647.0 9065.800825 72.390000 1.667 311623.799 south
3 Canada 33390141.0 36319.235010 80.653000 16.762 540431.495 north
4 Chile 16284741.0 13171.638850 78.553000 3.343 54434.634 south
5 Colombia 44227550.0 7006.580419 72.889000 1.238 53585.300 south
6 Costa Rica 4133884.0 9645.061420 78.782000 1.286 5463.059 south
7 Cuba 11416987.0 8948.102923 78.273000 2.220 25051.431 south
8 Dominican Republic 9319622.0 6025.374752 72.235000 1.897 17522.139 south
9 Ecuador 13755680.0 6873.262326 74.994000 1.742 23926.725 south
10 El Salvador 6939688.0 5728.353514 71.878000 1.037 6252.815 south
11 Guatemala 12572928.0 5186.050003 70.259000 0.811 10621.597 south
12 Haiti 8502814.0 1201.637154 60.916000 0.214 1980.992 south
13 Honduras 7483763.0 3548.330846 70.198000 0.976 7192.737 south
14 Jamaica 2780132.0 7320.880262 72.567000 3.746 10281.648 south
15 Mexico 108700891.0 11977.574960 76.195000 3.854 412385.135 north
16 Nicaragua 5675356.0 2749.320965 72.899000 0.750 4032.083 south
17 Panama 3242173.0 9809.185636 75.537000 2.035 6776.118 south
18 Paraguay 6667147.0 4172.838464 71.752000 0.599 3472.665 south
19 Peru 28674757.0 7408.905561 71.421000 1.037 28632.888 south
20 Trinidad and Tobago 1056608.0 18008.509240 69.819000 13.243 17175.823 south
21 United States 305082438.0 42646.380702 78.248513 19.268 5703220.175 north
22 Uruguay 3447496.0 10611.462990 76.384000 1.549 5151.871 south
23 Venezuela 26084662.0 11415.805690 73.747000 5.141 137701.548 south
Now we can use this column to repeat our groupby() method.
(
gapminder_co2
.assign(region=lambda df: np.where(df['country'].isin(['Canada', 'United States', 'Mexico']), 'north', 'south'))
.groupby('region')[["emissions_total", "pop"]]
.sum()
)
emissions_total pop
region
north 6656036.805 447173470.0
south 889331.721 451697714.0
We see that although Canada, the United States, and Mexico account for close to half the population of the Americas, they account for 88% of the CO2 emitted. We just did this math quickly by plugging the numbers from our table into the console to get the percentages. Can we make that a little more reproducible by calculating percentages for population and total emissions into our data before summarizing?
Map plots
The plotly library also has useful functions to draw your data on a map. There are lots of different ways to draw maps but here’s a quick example of making a choropleth map using the gapminder data. Here we will plot each country with a color indicating the life expectancy in 1997.
In order for the map function px.choropleth() to understand the countries in the gapminder data,
we need to first convert the country names to standard 3-letter country codes.
NOTE: we haven’t learned how to modify the data in this way yet, but we’ll learn about that in the next lesson. Just take for granted that it works for now :)
(
gapminder_1997
.replace({'country' : {'United States' : 'United States of America',
'United Kingdom' : 'United Kingdom of Great Britain and Northern Ireland',
}})
.merge(pd.read_csv("./data/country-iso.csv")
.rename(columns={'name' : 'country'}),
on='country', how='inner')
.pipe(px.choropleth,
locations='alpha-3',
color='lifeExp',
hover_name='country',
hover_data=['lifeExp', 'pop'])
)
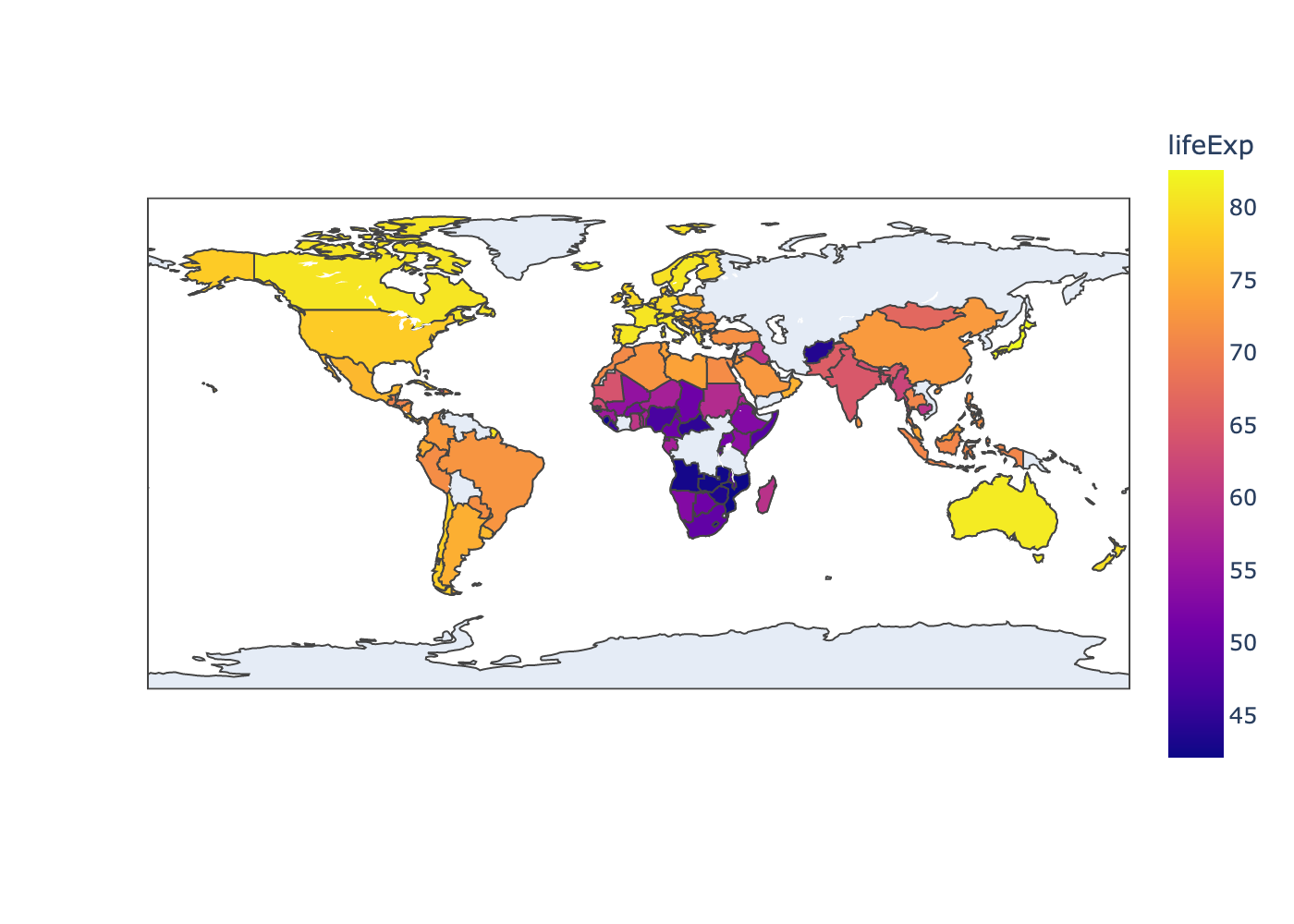
Notice that this map helps to show that we actually have some gaps in the data. We are missing observations for countries like Russia and many countries in central Africa. Thus, it’s important to acknowledge that any patterns or trends we see in the data might not apply to those regions.
Finishing with Git and GitHub
Awesome work! Let’s make sure it doesn’t go to waste. Time to add, commit, and push our changes to GitHub again - do you remember how?
changing directories
Print your current working directory and list the items in the directory to check where you are. If you are not in the un-report directory, navigate there.
Solution:
pwd ls cd ~/Desktop/un-report ls
reviewing git and GitHub
Pull to make sure our local repository is up to date. Then add, commit, and push your commits to GitHub. Don’t forget to check your git status periodically to make sure everything is going as expected!
Solution:
git status git pull git status git add "gapminder_data_analysis.ipynb" git status git commit -m "Create data analysis file" git status git log --online git push git status
Bonus exercises
Calculating percent
What percentage of the population and CO2 emissions in the Americas does the United States make up? What percentage of the population and CO2 emission does North America make up?
Solution
Create a new variable using
assign()that calculates percentages for the pop and total variables.( gapminder_co2 .assign(region=lambda df: np.where(df['country'].isin(['Canada', 'United States', 'Mexico']), 'north', 'south'), emissions_total_perc=lambda df: df['emissions_total']/df['emissions_total'].sum()*100, pop_perc=lambda df: df['pop']/df['pop'].sum()*100) )country pop gdpPercap lifeExp emissions_percap emissions_total region emissions_total_perc pop_perc 0 Argentina 40301927.0 12779.379640 75.320000 3.819 149476.040 south 1.981030 4.483615 1 Bolivia 9119152.0 3822.137084 65.554000 0.984 8975.809 south 0.118958 1.014512 2 Brazil 190010647.0 9065.800825 72.390000 1.667 311623.799 south 4.130001 21.138807 3 Canada 33390141.0 36319.235010 80.653000 16.762 540431.495 north 7.162427 3.714675 4 Chile 16284741.0 13171.638850 78.553000 3.343 54434.634 south 0.721431 1.811688 5 Colombia 44227550.0 7006.580419 72.889000 1.238 53585.300 south 0.710175 4.920344 6 Costa Rica 4133884.0 9645.061420 78.782000 1.286 5463.059 south 0.072403 0.459897 7 Cuba 11416987.0 8948.102923 78.273000 2.220 25051.431 south 0.332011 1.270147 8 Dominican Republic 9319622.0 6025.374752 72.235000 1.897 17522.139 south 0.232224 1.036814 9 Ecuador 13755680.0 6873.262326 74.994000 1.742 23926.725 south 0.317105 1.530328 10 El Salvador 6939688.0 5728.353514 71.878000 1.037 6252.815 south 0.082870 0.772045 11 Guatemala 12572928.0 5186.050003 70.259000 0.811 10621.597 south 0.140770 1.398746 12 Haiti 8502814.0 1201.637154 60.916000 0.214 1980.992 south 0.026254 0.945944 13 Honduras 7483763.0 3548.330846 70.198000 0.976 7192.737 south 0.095327 0.832573 14 Jamaica 2780132.0 7320.880262 72.567000 3.746 10281.648 south 0.136264 0.309291 15 Mexico 108700891.0 11977.574960 76.195000 3.854 412385.135 north 5.465407 12.093044 16 Nicaragua 5675356.0 2749.320965 72.899000 0.750 4032.083 south 0.053438 0.631387 17 Panama 3242173.0 9809.185636 75.537000 2.035 6776.118 south 0.089805 0.360694 18 Paraguay 6667147.0 4172.838464 71.752000 0.599 3472.665 south 0.046024 0.741724 19 Peru 28674757.0 7408.905561 71.421000 1.037 28632.888 south 0.379476 3.190085 20 Trinidad and Tobago 1056608.0 18008.509240 69.819000 13.243 17175.823 south 0.227634 0.117548 21 United States 305082438.0 42646.380702 78.248513 19.268 5703220.175 north 75.585707 33.940618 22 Uruguay 3447496.0 10611.462990 76.384000 1.549 5151.871 south 0.068279 0.383536 23 Venezuela 26084662.0 11415.805690 73.747000 5.141 137701.548 south 1.824981 2.901936This table shows that the United states makes up 33% of the population of the Americas, but accounts for 76% of total emissions. Now let’s take a look at population and emission for the two different continents:
( gapminder_co2 .assign(region=lambda df: np.where(df['country'].isin(['Canada', 'United > > States', 'Mexico']), 'north', 'south'), emissions_total_perc=lambda df: df['emissions_total']/df> > ['emissions_total'].sum()*100, pop_perc=lambda df: df['pop']/df['pop'].sum()*100) .groupby('region') .agg({'emissions_total_perc' : 'sum', 'pop_perc' : 'sum'}) )emissions_total_perc pop_perc region north 88.213542 49.748337 south 11.786458 50.251663
CO2 bar plot
Create a bar plot of the percent of emissions for each country, colored by north and south America.
Solution
( gapminder_co2 .assign(region=lambda df: np.where(df['country'].isin(['Canada', 'United > > States', 'Mexico']), 'north', 'south'), emissions_total_perc=lambda df: df['emissions_total']/df> > ['emissions_total'].sum()*100, pop_perc=lambda df: df['pop']/df['pop'].sum()*100) .pipe(so.Plot, x='country', y='emissions_total_perc', color='region') .add(so.Bar()) )
Now switch the x and y axis to make the country names more readable.
Solution
( gapminder_co2 .assign(region=lambda df: np.where(df['country'].isin(['Canada', 'United > > States', 'Mexico']), 'north', 'south'), emissions_total_perc=lambda df: df['emissions_total']/df> > ['emissions_total'].sum()*100, pop_perc=lambda df: df['pop']/df['pop'].sum()*100) .pipe(so.Plot, x='emissions_total_perc', y='country', color='region') .add(so.Bar()) )
Reorder the bars in descending order. Hint: what method we used earlier to sort data values?
Solution
( gapminder_co2 .assign(region=lambda df: np.where(df['country'].isin(['Canada', 'United > > States', 'Mexico']), 'north', 'south'), emissions_total_perc=lambda df: df['emissions_total']/df> > ['emissions_total'].sum()*100, pop_perc=lambda df: df['pop']/df['pop'].sum()*100) .sort_values('emissions_total_perc', ascending=False) .pipe(so.Plot, x='emissions_total_perc', y='country', color='region') .add(so.Bar()) )
Practice making it look pretty!
low emissions
Find the 3 countries with lowest per capita emissions.
Solution
( gapminder_co2 .assign(region=lambda df: np.where(df['country'].isin(['Canada', 'United States', 'Mexico']), 'north', 'south'), emissions_total_perc=lambda df: df['emissions_total']/df['emissions_total'].sum()*100, pop_perc=lambda df: df['pop']/df['pop'].sum()*100,) .sort_values('emissions_percap', ascending=True)[['country', 'emissions_percap']] .head(3) )country emissions_percap 12 Haiti 0.214 18 Paraguay 0.599 16 Nicaragua 0.750Create a bar chart for the per capita emissions for just those three countries.
Solution
( gapminder_co2 .assign(region=lambda df: np.where(df['country'].isin(['Canada', 'United States', 'Mexico']), 'north', 'south'), emissions_total_perc=lambda df: df['emissions_total']/df['emissions_total'].sum()*100, pop_perc=lambda df: df['pop']/df['pop'].sum()*100,) .query("country in ['Haiti', 'Paraguay', 'Nicaragua']") .pipe(so.Plot, x='country', y='emissions_percap') .add(so.Bar()) )
Reorder them in descending order.
Solution
( gapminder_co2 .assign(region=lambda df: np.where(df['country'].isin(['Canada', 'United States', 'Mexico']), 'north', 'south'), emissions_total_perc=lambda df: df['emissions_total']/df['emissions_total'].sum()*100, pop_perc=lambda df: df['pop']/df['pop'].sum()*100) .query("country in ['Haiti', 'Paraguay', 'Nicaragua']") .sort_values('emissions_percap', ascending=False) .pipe(so.Plot, x='country', y='emissions_percap') .add(so.Bar()) )
Key Points
Library importing is an important first step in preparing a Python environment.
Data analysis in Python facilitates reproducible research.
There are many useful methods in the pandas library that can aid in data analysis.
Assessing data source and structure is an important first step in analysis.
Preparing data for analysis can take significant effort and planning.